Gene
KWMTBOMO03495
Pre Gene Modal
BGIBMGA006347
Annotation
PREDICTED:_DNA_helicase_MCM9-like_isoform_X1_[Amyelois_transitella]
Full name
DNA helicase MCM9
Alternative Name
Mini-chromosome maintenance deficient domain-containing protein 1
Minichromosome maintenance 9
Minichromosome maintenance 9
Location in the cell
Nuclear Reliability : 3.974
Sequence
CDS
ATGATTTTAGAATATATATTAAAGTTCCATTTGGATGATTGTTTTAATGTATTATCAGAAAAAGATAATCTAGGATATTTTAGCATTAAAATCGATTTTTTAAAATTATTTGAAAACCATCCTGATGTGGGAGACAGAGTTCTGTGTGCTCCCATTGAAACCCTGCCCATATGCAATAAAGATATAGTGATGGCCCAACAACACATTATAGAATCTGAGGAATTTAAAAATACAGTTTCGAAATTAGAATATGTTTTCGTAAAGAAGAATGTTCATGCCAGATTTTATGGTTTACCTGTTTGTCCGGAGTTACACAGAACAGTTTTCCCTAAAAATGTAGATCTGGGATGTTTTTTAAAAGTAACAGGTACAGTTGTCCGAGTGACCCAGTCCAAAATGTTGGAATATCAAAGAAAGTATGTCTGTATGAAATGCAAATATGAGAATTATGTTCAAGCAGATTTCGAAAGGAGATACATTCTTAAATCACCATCAAAATGTTGTAATATTGATCCAAAATGCAGAAGTACAATATTCTCACAAGTGAATCTTGTATCCAGAGAATATTGTAAAGATTATCAGGAAATAAAAATTCAGGAGCAAGTTAACAAGCTTAGCATTGGTACCATTCCAGGCTCATTCTGGGTGGTTTTAGAAGATGATTTAGTAGATTGTTGTAAACCTGGTGATGATGTTATAATTTGCGGTACTATTAGAAGACGTTGGCGACCATCATTTCAAACAAAAAAATCTGAAGTGGAACTAGTCCTCCAAGCAAATTACATAGAAGTGTGTAATGCGCTAAGATCTAAAGTGGTAGCCACAGCTCCTGACATAAAAGAATGCTTTGACAGTTTCTGGTCTTACTATGATTCGTGTCCTTTGAAAGGCAGAGATCATATACTTAGATCGATTTGTCCTCAAGTCTACGGTCTACATTTGGTAAAGTTGGCTGTGTTGCTCACGGTGATTACCGGATCGAATCACATAGTCGAGGAAACAGATGAAAAGAATACATCCTTTGACGATAATCATCATACAAAAGTGCGGGGTCAGTGTCATCTCCTGTTAGTTGGAGATCCAGGCACTGGGAAGTCGCAACTGCTTCGTTCAGGAGCAGATCTGACTTCGCGTTCGGTGTTCACTTCGGGAGCTGGCAGTACGCGAGCCGGCCTCACGTGTGCTGCGCTCAGGGAAGACGGCGAGTGGCAGCTAGAAGCTGGAGCACTGGTTCTGTCGGACGGCGGCGTGTGCTGTATAGATGAGATCTCCCAGCTGAAGGAGCACGACCGTACCGCCATACACGAGGCGATGGAGCAACAGACAATCTCCATAGCCAAGGCAGGTATAGTCTGCAAATTGAACACACGTTGTGCTGTGATAGCAGCCTGCAATCCTAAGGGGAACTACGATTCAAATCAACCGCTCAGTATGAATTTAGCACTTGGAACGCCACTTTTGAGCCGATTTGATCTAATTTTCATTCTATTGGATACAAAAAATAAAGCTTGGGATAAACTTGTATCTTCTTATATATTGTTCGGTGATTCAAATTTAGAACAAAATAAAAAATATTGGAATCTCGAAAAATTGCAGATGTACATTAGTCTAGTGGGACCAAGGACGACAAAGATGACAAAATCGGCAAACATTATCCTGCAATCGTATTATATGGCCCAGAGAAAAGCCGAAAGCAGAGATCCATCACGTACTACCGTGAGAATGCTCGACAGTCTGGTCAGACTATCACAAGCTCACTGCCGTTTGATGTATAGAACAACAATATATCCGATGGACGCCATTATAGCCGTATCTTTAGTAGATTTATCAATGCAAGACTGCACTTTATCAGACACTGCGGATGCGCTACATTCCAATTTCTATAAATACCCTGACTTTGAATACCTATGCACAGCAAAAAAACTATTAACAAAATTAAATTTGTACGAAATATGGCGAAATGAATTATTACACTACGGTAAATTACTTCAGGTCGATCACAAAACATTGGAACATGACATTGATAGCGGAATCATTAAGCTTTTCGCGAAATATGATGATGTCGCCGATGAAAATACACCTTTGTCAGCATCGGTAATAACAAGTTCTTATTTTAAGAAAGAAGAAAATGGATTGTTACATAATGACAAAAACAGTTTACCCGATGAAGAGAACAAAGATATCATAGAAGTAAATAAAATGTTGGCAGTGACATTGAAAAAACACGCAACTTTAAATAGAGACGAAAGAAAACTACCAGTAGCTCAGAAAAAACCTCAAAATAAAAGAAAACGGAAAGAAGTTACTGTGAATTCAAAAATTCTTAAAAGTTTAGTTCCTAAAAAGCGTAAGAAATCAAAGGAGGAAGTAAGGGATGCCAGTAGTCCTACAAACATTGACGTTGACCAGAATCATCTCTTAAATGCAGTGCCTAGTGTTAATGATGTGTTTGCAGAGTACGGTATCAATCTTAAGTTTGAAGGTGAAAGTCAACATAAATCAAGTGGTAAAAGAATCGAATTAAAAGAAAATAAGATTGATAATGAAATCATTGTTCCTAGAATTGAAAAAAAGGATCTTGTAGAAAACAAAACTAATTCTCATAAAGAACAAAATGCAAATATCTCAACAATAAATAAATTAAAACAGTTCGCATGTGAACAAAAATATAACATTGATAAATATGAAGAAAAGTTATCGATATCTGAATACAACAACAGTTTTAAATTTAATATTGAGAGTAAAAGTACAAATGCAGTTCAAGATAAGTCTGAACGAAGTTCCCAAATATCAATTTTTGAAAGTTCAGATTGTGATATTGATTTAGATTTATGA
Protein
MILEYILKFHLDDCFNVLSEKDNLGYFSIKIDFLKLFENHPDVGDRVLCAPIETLPICNKDIVMAQQHIIESEEFKNTVSKLEYVFVKKNVHARFYGLPVCPELHRTVFPKNVDLGCFLKVTGTVVRVTQSKMLEYQRKYVCMKCKYENYVQADFERRYILKSPSKCCNIDPKCRSTIFSQVNLVSREYCKDYQEIKIQEQVNKLSIGTIPGSFWVVLEDDLVDCCKPGDDVIICGTIRRRWRPSFQTKKSEVELVLQANYIEVCNALRSKVVATAPDIKECFDSFWSYYDSCPLKGRDHILRSICPQVYGLHLVKLAVLLTVITGSNHIVEETDEKNTSFDDNHHTKVRGQCHLLLVGDPGTGKSQLLRSGADLTSRSVFTSGAGSTRAGLTCAALREDGEWQLEAGALVLSDGGVCCIDEISQLKEHDRTAIHEAMEQQTISIAKAGIVCKLNTRCAVIAACNPKGNYDSNQPLSMNLALGTPLLSRFDLIFILLDTKNKAWDKLVSSYILFGDSNLEQNKKYWNLEKLQMYISLVGPRTTKMTKSANIILQSYYMAQRKAESRDPSRTTVRMLDSLVRLSQAHCRLMYRTTIYPMDAIIAVSLVDLSMQDCTLSDTADALHSNFYKYPDFEYLCTAKKLLTKLNLYEIWRNELLHYGKLLQVDHKTLEHDIDSGIIKLFAKYDDVADENTPLSASVITSSYFKKEENGLLHNDKNSLPDEENKDIIEVNKMLAVTLKKHATLNRDERKLPVAQKKPQNKRKRKEVTVNSKILKSLVPKKRKKSKEEVRDASSPTNIDVDQNHLLNAVPSVNDVFAEYGINLKFEGESQHKSSGKRIELKENKIDNEIIVPRIEKKDLVENKTNSHKEQNANISTINKLKQFACEQKYNIDKYEEKLSISEYNNSFKFNIESKSTNAVQDKSERSSQISIFESSDCDIDLDL
Summary
Description
Component of the MCM8-MCM9 complex, a complex involved in the repair of double-stranded DNA breaks (DBSs) and DNA interstrand cross-links (ICLs) by homologous recombination (HR) (PubMed:23401855, PubMed:22771120). Required for DNA resection by the MRE11-RAD50-NBN/NBS1 (MRN) complex at double-stranded DNA breaks to generate ssDNA by recruiting the MRN complex to the repair site and by promoting the complex nuclease activity (By similarity). Probably by regulating the localization of the MNR complex, indirectly regulates the recruitment of downstream effector RAD51 to DNA damage sites including DBSs and ICLs (PubMed:22771120, PubMed:23401855). Acts as a helicase in DNA mismatch repair (MMR) following DNA replication errors to unwind the mismatch containing DNA strand (PubMed:22771120, PubMed:26300262). In addition, recruits MLH1, a component of the MMR complex, to chromatin (By similarity). The MCM8-MCM9 complex is dispensable for DNA replication and S phase progression (PubMed:21987787). Probably by regulating HR, plays a key role during gametogenesis (PubMed:21987787, PubMed:22771120).
Component of the MCM8-MCM9 complex, a complex involved in the repair of double-stranded DNA breaks (DBSs) and DNA interstrand cross-links (ICLs) by homologous recombination (HR) (PubMed:23401855). Required for DNA resection by the MRE11-RAD50-NBN/NBS1 (MRN) complex by recruiting the MRN complex to the repair site and by promoting the complex nuclease activity (PubMed:26215093). Probably by regulating the localization of the MRN complex, indirectly regulates the recruitment of downstream effector RAD51 to DNA damage sites including DBSs and ICLs (PubMed:23401855). Acts as a helicase in DNA mismatch repair (MMR) following DNA replication errors to unwind the mismatch containing DNA strand (PubMed:26300262). In addition, recruits MLH1, a component of the MMR complex, to chromatin (PubMed:26300262). The MCM8-MCM9 complex is dispensable for DNA replication and S phase progression (PubMed:23401855). Probably by regulating HR, plays a key role during gametogenesis (By similarity).
Component of the MCM8-MCM9 complex, a complex involved in homologous recombination repair following DNA interstrand cross-links and plays a key role during gametogenesis. The MCM8-MCM9 complex probably acts as a hexameric helicase required to process aberrant forks into homologous recombination substrates and to orchestrate homologous recombination with resection, fork stabilization and fork restart. In eggs, required for MCM2-7 loading onto chromatin during DNA replication. Probably not required for DNA replication in other cells (By similarity).
Component of the MCM8-MCM9 complex, a complex involved in the repair of double-stranded DNA breaks (DBSs) and DNA interstrand cross-links (ICLs) by homologous recombination (HR) (PubMed:23401855). Required for DNA resection by the MRE11-RAD50-NBN/NBS1 (MRN) complex by recruiting the MRN complex to the repair site and by promoting the complex nuclease activity (PubMed:26215093). Probably by regulating the localization of the MRN complex, indirectly regulates the recruitment of downstream effector RAD51 to DNA damage sites including DBSs and ICLs (PubMed:23401855). Acts as a helicase in DNA mismatch repair (MMR) following DNA replication errors to unwind the mismatch containing DNA strand (PubMed:26300262). In addition, recruits MLH1, a component of the MMR complex, to chromatin (PubMed:26300262). The MCM8-MCM9 complex is dispensable for DNA replication and S phase progression (PubMed:23401855). Probably by regulating HR, plays a key role during gametogenesis (By similarity).
Component of the MCM8-MCM9 complex, a complex involved in homologous recombination repair following DNA interstrand cross-links and plays a key role during gametogenesis. The MCM8-MCM9 complex probably acts as a hexameric helicase required to process aberrant forks into homologous recombination substrates and to orchestrate homologous recombination with resection, fork stabilization and fork restart. In eggs, required for MCM2-7 loading onto chromatin during DNA replication. Probably not required for DNA replication in other cells (By similarity).
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Subunit
Component of the MCM8-MCM9 complex, which forms a hexamer composed of MCM8 and MCM9 (PubMed:22771120). Interacts with the DNA mismatch repair (MMR) complex composed at least of MSH2, MSH3, MSH6, PMS1 and MLH1 (By similarity). Interacts with MLH1; the interaction recruits MLH1 to chromatin (By similarity). Interacts with MSH2; the interaction recruits MCM9 to chromatin (By similarity). Interacts with MSH6 (By similarity). Interacts with the MRN complex composed of MRE11, RAD50 and NBN/NBS1; the interaction recruits the MRN complex to DNA damage sites (By similarity). Interacts with RAD51; the interaction recruits RAD51 to DNA damage sites (By similarity).
Component of the MCM8-MCM9 complex, which forms a hexamer composed of MCM8 and MCM9 (PubMed:23401855, PubMed:26300262, PubMed:26215093). Interacts with the DNA mismatch repair (MMR) complex composed at least of MSH2, MSH3, MSH6, PMS1 and MLH1 (PubMed:26300262). Interacts with MLH1; the interaction recruits MLH1 to chromatin (PubMed:26300262). Interacts with MSH2; the interaction recruits MCM9 to chromatin (PubMed:26300262). Interacts with MSH6 (PubMed:26300262). Interacts with the MRN complex composed of MRE11, RAD50 and NBN/NBS1; the interaction recruits the MRN complex to DNA damage sites (PubMed:26215093). Interacts with RAD51; the interaction recruits RAD51 to DNA damage sites (PubMed:23401855).
Component of the MCM8-MCM9 complex, which forms a hexamer composed of mcm8 and mcm9. Interacts with cdt1 (By similarity).
Component of the MCM8-MCM9 complex, which forms a hexamer composed of MCM8 and MCM9 (PubMed:23401855, PubMed:26300262, PubMed:26215093). Interacts with the DNA mismatch repair (MMR) complex composed at least of MSH2, MSH3, MSH6, PMS1 and MLH1 (PubMed:26300262). Interacts with MLH1; the interaction recruits MLH1 to chromatin (PubMed:26300262). Interacts with MSH2; the interaction recruits MCM9 to chromatin (PubMed:26300262). Interacts with MSH6 (PubMed:26300262). Interacts with the MRN complex composed of MRE11, RAD50 and NBN/NBS1; the interaction recruits the MRN complex to DNA damage sites (PubMed:26215093). Interacts with RAD51; the interaction recruits RAD51 to DNA damage sites (PubMed:23401855).
Component of the MCM8-MCM9 complex, which forms a hexamer composed of mcm8 and mcm9. Interacts with cdt1 (By similarity).
Similarity
Belongs to the MCM family.
Keywords
Alternative splicing
ATP-binding
Chromosome
Complete proteome
DNA damage
DNA repair
DNA-binding
Helicase
Hydrolase
Nucleotide-binding
Nucleus
Phosphoprotein
Reference proteome
DNA replication
Feature
chain DNA helicase MCM9
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A2A4JLZ1
A0A2W1BW94
A0A194QBS7
A0A212EIY5
A0A194QXS1
A0A067RI06
+ More
A0A1B6C633 K1PZI7 A0A210PG05 A0A1S3GJ75 V8PAR1 A0A2Y9HI90 A0A2K6T8N1 A0A2R8ZWC4 A0A093I0Q0 A0A384AUU8 A0A2J8TAG6 Q2KHI9 Q9NXL9 Q2KHI9-2 G3U4T6 A0A1W4ZB32 G3TC88 F6RIX4 A0A3B3SWV2 A0A340X4S7 W4XJQ6 A0A2G8KKN5 A0A3N0Z2H5 D6WB76 A0A146MDY4 W5LF76 A0A0A9XC16 M3XF20 A0A2C9JZD6
A0A1B6C633 K1PZI7 A0A210PG05 A0A1S3GJ75 V8PAR1 A0A2Y9HI90 A0A2K6T8N1 A0A2R8ZWC4 A0A093I0Q0 A0A384AUU8 A0A2J8TAG6 Q2KHI9 Q9NXL9 Q2KHI9-2 G3U4T6 A0A1W4ZB32 G3TC88 F6RIX4 A0A3B3SWV2 A0A340X4S7 W4XJQ6 A0A2G8KKN5 A0A3N0Z2H5 D6WB76 A0A146MDY4 W5LF76 A0A0A9XC16 M3XF20 A0A2C9JZD6
EC Number
3.6.4.12
Pubmed
28756777
26354079
22118469
24845553
22992520
28812685
+ More
24297900 22722832 19468303 15489334 16141072 16226853 15850810 21987787 22771120 23401855 26300262 14702039 14574404 18669648 20068231 23403237 23186163 25480036 26215093 20431018 29240929 29023486 18362917 19820115 26823975 25329095 25401762 17975172 15562597
24297900 22722832 19468303 15489334 16141072 16226853 15850810 21987787 22771120 23401855 26300262 14702039 14574404 18669648 20068231 23403237 23186163 25480036 26215093 20431018 29240929 29023486 18362917 19820115 26823975 25329095 25401762 17975172 15562597
EMBL
NWSH01001141
PCG72422.1
KZ149899
PZC78611.1
KQ459249
KPJ02445.1
+ More
AGBW02014560 OWR41431.1 KQ461155 KPJ08376.1 KK852460 KDR23496.1 GEDC01028335 JAS08963.1 JH819194 EKC29607.1 NEDP02076730 OWF35387.1 AZIM01000436 ETE71083.1 AJFE02097141 AJFE02097142 AJFE02097143 AJFE02097144 KL206669 KFV84797.1 NDHI03003512 PNJ30010.1 PNJ30014.1 AC153949 AC155941 BC062185 AK018494 AK046636 BN000883 AK299076 AK000177 AL132874 AL359634 BC031658 BN000882 AAMC01008549 AAGJ04116641 MRZV01000515 PIK48566.1 RJVU01015592 ROL52501.1 KQ971312 EEZ97925.1 GDHC01000621 JAQ18008.1 GBHO01026428 JAG17176.1 AANG04000169
AGBW02014560 OWR41431.1 KQ461155 KPJ08376.1 KK852460 KDR23496.1 GEDC01028335 JAS08963.1 JH819194 EKC29607.1 NEDP02076730 OWF35387.1 AZIM01000436 ETE71083.1 AJFE02097141 AJFE02097142 AJFE02097143 AJFE02097144 KL206669 KFV84797.1 NDHI03003512 PNJ30010.1 PNJ30014.1 AC153949 AC155941 BC062185 AK018494 AK046636 BN000883 AK299076 AK000177 AL132874 AL359634 BC031658 BN000882 AAMC01008549 AAGJ04116641 MRZV01000515 PIK48566.1 RJVU01015592 ROL52501.1 KQ971312 EEZ97925.1 GDHC01000621 JAQ18008.1 GBHO01026428 JAG17176.1 AANG04000169
Proteomes
UP000218220
UP000053268
UP000007151
UP000053240
UP000027135
UP000005408
+ More
UP000242188 UP000081671 UP000248481 UP000233220 UP000240080 UP000053584 UP000000589 UP000005640 UP000007646 UP000192224 UP000008143 UP000261540 UP000265300 UP000007110 UP000230750 UP000007266 UP000018467 UP000011712 UP000076420
UP000242188 UP000081671 UP000248481 UP000233220 UP000240080 UP000053584 UP000000589 UP000005640 UP000007646 UP000192224 UP000008143 UP000261540 UP000265300 UP000007110 UP000230750 UP000007266 UP000018467 UP000011712 UP000076420
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JLZ1
A0A2W1BW94
A0A194QBS7
A0A212EIY5
A0A194QXS1
A0A067RI06
+ More
A0A1B6C633 K1PZI7 A0A210PG05 A0A1S3GJ75 V8PAR1 A0A2Y9HI90 A0A2K6T8N1 A0A2R8ZWC4 A0A093I0Q0 A0A384AUU8 A0A2J8TAG6 Q2KHI9 Q9NXL9 Q2KHI9-2 G3U4T6 A0A1W4ZB32 G3TC88 F6RIX4 A0A3B3SWV2 A0A340X4S7 W4XJQ6 A0A2G8KKN5 A0A3N0Z2H5 D6WB76 A0A146MDY4 W5LF76 A0A0A9XC16 M3XF20 A0A2C9JZD6
A0A1B6C633 K1PZI7 A0A210PG05 A0A1S3GJ75 V8PAR1 A0A2Y9HI90 A0A2K6T8N1 A0A2R8ZWC4 A0A093I0Q0 A0A384AUU8 A0A2J8TAG6 Q2KHI9 Q9NXL9 Q2KHI9-2 G3U4T6 A0A1W4ZB32 G3TC88 F6RIX4 A0A3B3SWV2 A0A340X4S7 W4XJQ6 A0A2G8KKN5 A0A3N0Z2H5 D6WB76 A0A146MDY4 W5LF76 A0A0A9XC16 M3XF20 A0A2C9JZD6
PDB
3F9V
E-value=2.13819e-77,
Score=739
Ontologies
GO
GO:0003677
GO:0006270
GO:0005524
GO:0008380
GO:0005681
GO:0004386
GO:0097362
GO:0032406
GO:0032408
GO:0000724
GO:0032407
GO:0070716
GO:0005634
GO:0007292
GO:0003678
GO:0019899
GO:0036298
GO:0071168
GO:0003682
GO:0032508
GO:0006974
GO:0007276
GO:0044877
GO:0003688
GO:0042555
GO:0003697
GO:0005694
GO:0006260
GO:0003676
GO:0004352
GO:0000287
GO:0003723
GO:0048384
PANTHER
Topology
Subcellular location
Nucleus
Colocalizes to nuclear foci with RPA1 following DNA damage (PubMed:23401855). Localizes to double-stranded DNA breaks (By similarity). Recruited to chromatin by MSH2 (By similarity). With evidence from 1 publications.
Chromosome Colocalizes to nuclear foci with RPA1 following DNA damage (PubMed:23401855). Localizes to double-stranded DNA breaks (By similarity). Recruited to chromatin by MSH2 (By similarity). With evidence from 1 publications.
Chromosome Colocalizes to nuclear foci with RPA1 following DNA damage (PubMed:23401855). Localizes to double-stranded DNA breaks (By similarity). Recruited to chromatin by MSH2 (By similarity). With evidence from 1 publications.
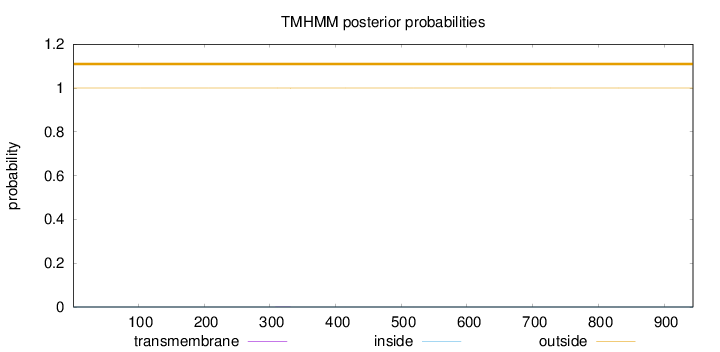
Length:
944
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0087
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00028
outside
1 - 944
Population Genetic Test Statistics
Pi
203.925675
Theta
195.770812
Tajima's D
-1.796531
CLR
128.875481
CSRT
0.0284985750712464
Interpretation
Uncertain
