Gene
KWMTBOMO03491
Pre Gene Modal
BGIBMGA006348
Annotation
Robo1a_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.744 Nuclear Reliability : 1.388 PlasmaMembrane Reliability : 1.057
Sequence
CDS
ATGATGGATGCTGGTGTTAATGAGTGCGATAACGCGGCAAATGGAAGGAACCAATTCGTGTTTCTTGTGCTTGTGCATTTACTACTTATCGGACTAAATGGGACTAATGCACAGAATAGAGCACCTCGCATCAAAGAGCATCCGTCGAATACTGTGTCAGGACGCAGTGAACCGGCAACATTGAGATGTGTCGTGGAAGGCCGACCTAAACCTTATGTACAGTGGTTCAAAGACGGATTACCCTTGCCTCAAGCAGAAGACGGACACAGGGTCTTACTCGAAGACGGGCTGTTATTCTTGAGGGTCAACCGAGGGAAGAAAGATAGCGATGAGGGTATTTATTGGTGTGTTGCACGCAACATTGCCGGTGAGGCCGTCAGCCAGAATGCTTCGCTTAATGTGGCTATGCTAAGAGATGAATTCCGAATGGAGCCTAGGGATGTGCAAGTGGCCGCTGGTGAGCCAGCACTATTAGAGTGTTCTCCTCCAAGAGGAGTACCAGAACCATCTGTGCAATGGCTTAAAGACGGACAATCCTACGATGTAGAAGTTAATGGAAGAGTCACAATAACAGATACAGGAAACCTTAAAATTTTAGAAACACTACCCACTGATAGTGGTTTATTTCGATGCGTGGCGACAAATATAGCCGGCGAACGACAGTCAAGAGCCGCTGCTTTAATTGTTTTAAGACGACCACATTTCGTTGTTAAACCTAGTAATGTTACTGCTTTAGTGGGTCAAACTGTCGAATTTAACTGCCAGAGTTACGACTCGAACGTAAAAATAACTTGGGTCAAGGAAGATGGAACGCTGCCGTCTCATGCAACAATAATTCGTGGACTACTAAAATTAGAGCACGTATCTGCAACTGATGCTGGCGTCTATTCATGTCGTGCAGACAGCCACGCGGGAACCAGTATCACCAGTGCCACACTCACGGTTTATTCATTACCGCATTTCACGCAGATACCGTCCAACCTAACAGCTTGGGAAGGCGATGTTGTATCTATTCCATGCGATGCCAAAGGATTACCAACGCCGACGACGTTTTGGATATTAGAAGGCAAACAAGATTCGATGCTTTTTCCGGAATGTACTAGAAGCAATACGAGTTTGTTGTATTTGGACAGTGCGCGGGCGGAGCAAAGTGGAAGGGTAATTTGTACTGCAGTTAACGCAGCTGGAAGTATCATGCAAGAAGCGTATCTTACCGTTCTTAGAAAAGGAAATAGCGAGTTAAGAGACCATACGGATGATGACTTCGAAGATGATTTAGATCGCAATGTTCATATGACCGAGTATGAACTTCTTCAAGCGAGAAAATATCTCCAGCAAAACGTATTAGTGTTGAAGCGAATTGAAATGCTATCATCTTCAAGTGTTAAAGTTGTTTGGGATGTGCTAACAGACTATAATGAATATTTAGAAGGGGTTAAAATATGGTACAATGGGACAACGTTAAATTGGCGAAATATCACAGAAGAAGATTTAACTCAACAAAATTCACATTCAAACACTATTCTGTGCTTAAATGGATCTCTCACCAATGTTGACAACAGTGGTTCATCAAGCTATATACTGTCAGGATTGATGACTTACACGCAATATGATATATTTTTGATGCCATTTTATAAGACCCTTCTAGGGAAGCCTTCCAACATGATGAGTGGGTTTACGGATGAGGGTGTACCGTCGGCACCACCCCAAAGCGTCACAGCAGGGGTGATAAACGCAACATCGGCGTGGATACGGTGGGAGCCACCACCTGTGAGGACTTGGAACGGCGAACTTACCGGTTATTTGATAGAGATACGCGTGAGTGGCAGCACAGGTGGTCGGGTTGTGGGTCAGATGTCATTGAACTCCCGCACCCGCGCCGCAGCCGCGAGCTCCCTCCGCGCGGGTGGCCGCTACACCGCGCGGGCGGCCGCCGTCACGCGTCGCGGACACGGCCCATTCAGTGCACCGGCGCTGATACACATGCTGCCAACTCAGTCGCAGAGGCATTACGTACAGACTGAACCACCAAAAGATTCAAATATACCGAACATACTACAAGAGACTTGGCTCTTGGCTCTCGGGCTCACCCTTTTCTCTGTCATCATCATTGGAGCCGTCAGCATTTACTACATCAAACGGAGAAACAACATACAAAGAAAAAAGTCTGCCGGTCAATCGATTGTCACGGCTCAACAATGTCTCCTAAATAAAGACACGGTTTGGCTTCGCGACAGGCCCGTATTCGGACCGCCTAACGACACGAGCATCGACGTCAACAGCTGTCACCAGAGTCTTTTACATAATGGACAAACCACAAGTATTCTGAACGTGGAACCAGAGTATAGCTTACCGCAGAACTCCATTCAAGGACTAGACGCATCAAGCGTCGACCGTATCAGACGCGGACCACCGGAGCCATACGCGTCAAGCGCCATTTACACAGAACTCAATTTTAAGGACAACAACGAAATCGGAGACCTGAGAGGCTGTCCGGACAGACGATCTCATAACAATAGCATTGTAAGATCGTGTAATGGAAGCATACAATATTCAAATGGCGAATGCTCGACTTGTTGTCACAGTACTTCGAGTAGATCGACCAAAGACTACAGAGAGCTAAGACACGACAATGTACAGAATGAAATTAATGTCTTGGAGGACGACTGCGGTTCTTGCCACACGAACAGATCTGGATCGAGGAGTAGACAGGACAAAAGCAAAGTTTGTCCGGCCGCTGATTTAGATTACGACTATCCCCAATGGCATTGGCTGGGACGGGAGAACAGCTTCAAAATTCCCATCCAAACGAGCAGACATTCGAACATCGCGAGGTCGGAGCAGGGCTGTCAAATAAATCTATGTGATATACTGCCGCCGCCGCCGTATGAGAGTCCAGAAAACAGGCACAACTATGGATCGGAAAAGAATTAG
Protein
MMDAGVNECDNAANGRNQFVFLVLVHLLLIGLNGTNAQNRAPRIKEHPSNTVSGRSEPATLRCVVEGRPKPYVQWFKDGLPLPQAEDGHRVLLEDGLLFLRVNRGKKDSDEGIYWCVARNIAGEAVSQNASLNVAMLRDEFRMEPRDVQVAAGEPALLECSPPRGVPEPSVQWLKDGQSYDVEVNGRVTITDTGNLKILETLPTDSGLFRCVATNIAGERQSRAAALIVLRRPHFVVKPSNVTALVGQTVEFNCQSYDSNVKITWVKEDGTLPSHATIIRGLLKLEHVSATDAGVYSCRADSHAGTSITSATLTVYSLPHFTQIPSNLTAWEGDVVSIPCDAKGLPTPTTFWILEGKQDSMLFPECTRSNTSLLYLDSARAEQSGRVICTAVNAAGSIMQEAYLTVLRKGNSELRDHTDDDFEDDLDRNVHMTEYELLQARKYLQQNVLVLKRIEMLSSSSVKVVWDVLTDYNEYLEGVKIWYNGTTLNWRNITEEDLTQQNSHSNTILCLNGSLTNVDNSGSSSYILSGLMTYTQYDIFLMPFYKTLLGKPSNMMSGFTDEGVPSAPPQSVTAGVINATSAWIRWEPPPVRTWNGELTGYLIEIRVSGSTGGRVVGQMSLNSRTRAAAASSLRAGGRYTARAAAVTRRGHGPFSAPALIHMLPTQSQRHYVQTEPPKDSNIPNILQETWLLALGLTLFSVIIIGAVSIYYIKRRNNIQRKKSAGQSIVTAQQCLLNKDTVWLRDRPVFGPPNDTSIDVNSCHQSLLHNGQTTSILNVEPEYSLPQNSIQGLDASSVDRIRRGPPEPYASSAIYTELNFKDNNEIGDLRGCPDRRSHNNSIVRSCNGSIQYSNGECSTCCHSTSSRSTKDYRELRHDNVQNEINVLEDDCGSCHTNRSGSRSRQDKSKVCPAADLDYDYPQWHWLGRENSFKIPIQTSRHSNIARSEQGCQINLCDILPPPPYESPENRHNYGSEKN
Summary
Uniprot
EMBL
Proteomes
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
PDB
5OPE
E-value=2.16676e-80,
Score=765
Ontologies
GO
Topology
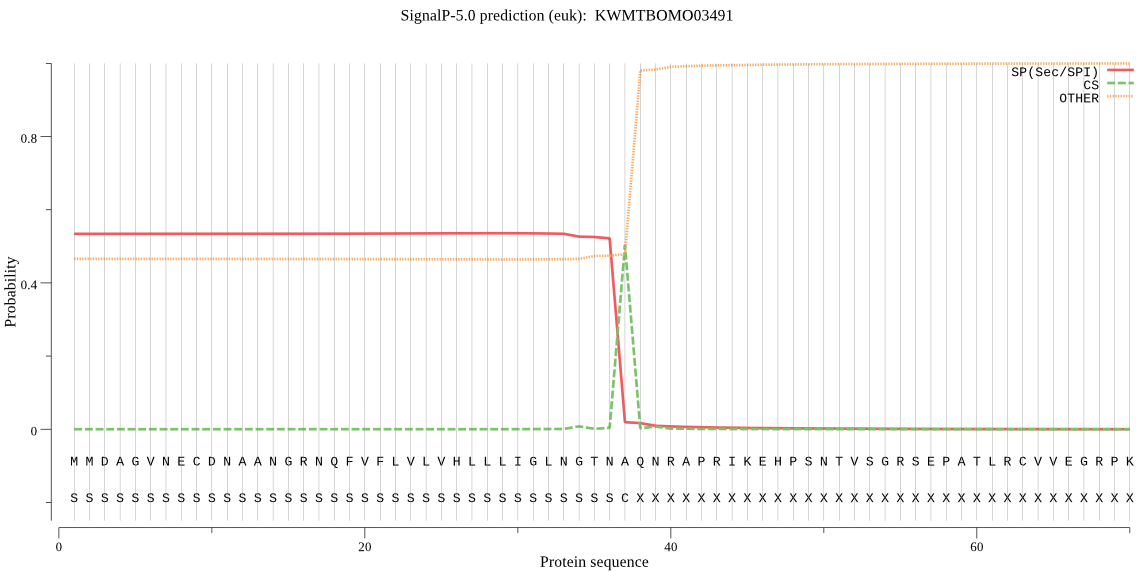
SignalP
Position: 1 - 37,
Likelihood: 0.536555
Length:
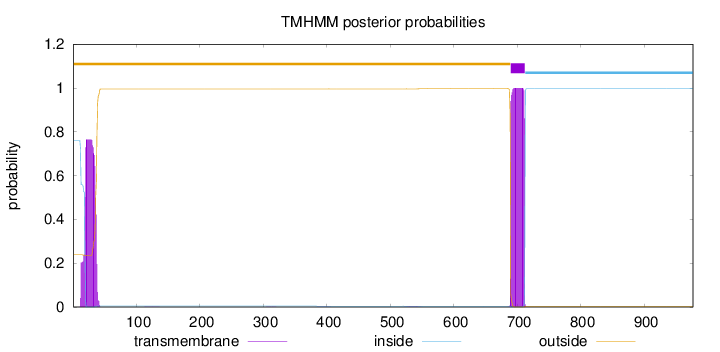
977
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
37.89648
Exp number, first 60 AAs:
14.96622
Total prob of N-in:
0.76003
POSSIBLE N-term signal
sequence
outside
1 - 689
TMhelix
690 - 712
inside
713 - 977
Population Genetic Test Statistics
Pi
244.289437
Theta
174.945345
Tajima's D
1.144796
CLR
0.001409
CSRT
0.700514974251287
Interpretation
Uncertain
