Gene
KWMTBOMO03487
Annotation
PREDICTED:_protein_held_out_wings_isoform_X3_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.514 Nuclear Reliability : 1.102
Sequence
CDS
ATGGTGACTACGCTCACAGAAAAAGTGTATGTGCCAGTCAAAGAACATCCCGATTTCAATTTCGTGGGAAGGATTCTAGGACCCAGAGGTATGACCGCAAAACAGCTGGAACAAGAAACAGGGTGCAAAATTATGGTCAGAGGAAAAGGGTCGATGAGGGACAAGAAAAAGGAAGATGCAAACAGGGGCAAACCCAATTGGGAACACCTAGCAGATGACCTCCACGTTCTGCTGACAGTTGAAGATACTGAAAACAGAGCCAAGATAAAGCTAGCGAGGGCCGTTGATGAAGTGAAGCGTCTCCTTGTGCCACAAGCTGACGGTGAAGATGAACTGAAGAAGCGCCAGTTGATGGAGTTGGCGATCATAAATGGAACTTACCGTGATTCCAGCTCGAAGGTGTCCGTTCCTGTGAACGGTCAGTGCCTAATAACTGACGAGGAATGGCGGCGCGTTGCAGCGGCGGCAGCCGAGACTCAGCGTTTGCTGGCACCAGGGGGCGTGGGCGGGGTGGCCCTGCGCACTCCCGCGCCCCTCGGCGCGCCCCTCATCCTGTCCCCTCGCATGTCCGCGCCAGCTCAACCCGGCCTTCTCAACGGGGCAGGGGCGGGAGCGGCGGCCGGCCTGCTGTCTCCGGGCGAGCCCGCGCATCAGCTGATCTACGCGTCGCCCTACGACTACGCGAACTACGCAGCTCTCACCGCGGCCGCGAACCCTCTGCTCGCCACGGAGTACGCCGCTGCTGATCATGCCGGTGTGGTGCGGAGGGCGGGGCGAGTCGCGATGAGGGAGCACCCCTACCAACGCACGGCCTTACCGGCGCCTTAA
Protein
MVTTLTEKVYVPVKEHPDFNFVGRILGPRGMTAKQLEQETGCKIMVRGKGSMRDKKKEDANRGKPNWEHLADDLHVLLTVEDTENRAKIKLARAVDEVKRLLVPQADGEDELKKRQLMELAIINGTYRDSSSKVSVPVNGQCLITDEEWRRVAAAAAETQRLLAPGGVGGVALRTPAPLGAPLILSPRMSAPAQPGLLNGAGAGAAAGLLSPGEPAHQLIYASPYDYANYAALTAAANPLLATEYAAADHAGVVRRAGRVAMREHPYQRTALPAP
Summary
Uniprot
A0A2W1C369
A0A2A4JN60
A0A2A4JLI9
A0A194RQ63
S4PLE9
A0A2A4JLZ6
+ More
A0A2A4JN30 A0A2A4JN54 A0A212EIX1 A0A2H1WV98 A0A1W4WTR5 N6UAX0 A0A194Q109 A0A0L7LQX0 A0A1W4WTR2 A0A1L8DGX2 B4NHR7 U5ERZ3 B4K6L5 A0A0L7QJT4 A0A0Q9XCC9 A0A0Q9WVE0 A0A3B0K9Q1 B4JRM0 B4M0M8 A0A0Q9X1V2 B4G314 A0A0K8V329 A0A1B6MTS9 A0A034VRQ6 A0A0K8U0M7 A0A0K8UR47 B3P7N6 A0A0P8ZYV7 A0A0Q9XBX3 A0A0Q9WFQ1 Q297U1 A0A0R3NHA9 A0A0P8YLR8 B3LVL6 A0A0R3NHJ4 A0A0P9C8K6 A0A0Q9WF42 A0A0P8XYV5 A0A0P8XYF7 A0A0R3NN08 A0A182PVU3 A0A1J1J5M5 A0A182U5H2 A0A182I7I4 A0A182LS51 A0A182VNH4 A0A182WXM0 A0A182RZB5 A0A182LMK0 A0A182WGB8 A0A146LT23 A0A2M4AL17 A0A182QCU5 A0A1S4GZS9 A0A084WQK6
A0A2A4JN30 A0A2A4JN54 A0A212EIX1 A0A2H1WV98 A0A1W4WTR5 N6UAX0 A0A194Q109 A0A0L7LQX0 A0A1W4WTR2 A0A1L8DGX2 B4NHR7 U5ERZ3 B4K6L5 A0A0L7QJT4 A0A0Q9XCC9 A0A0Q9WVE0 A0A3B0K9Q1 B4JRM0 B4M0M8 A0A0Q9X1V2 B4G314 A0A0K8V329 A0A1B6MTS9 A0A034VRQ6 A0A0K8U0M7 A0A0K8UR47 B3P7N6 A0A0P8ZYV7 A0A0Q9XBX3 A0A0Q9WFQ1 Q297U1 A0A0R3NHA9 A0A0P8YLR8 B3LVL6 A0A0R3NHJ4 A0A0P9C8K6 A0A0Q9WF42 A0A0P8XYV5 A0A0P8XYF7 A0A0R3NN08 A0A182PVU3 A0A1J1J5M5 A0A182U5H2 A0A182I7I4 A0A182LS51 A0A182VNH4 A0A182WXM0 A0A182RZB5 A0A182LMK0 A0A182WGB8 A0A146LT23 A0A2M4AL17 A0A182QCU5 A0A1S4GZS9 A0A084WQK6
Pubmed
EMBL
KZ149899
PZC78603.1
NWSH01001063
PCG72840.1
PCG72841.1
KQ459896
+ More
KPJ19460.1 GAIX01000626 JAA91934.1 PCG72839.1 PCG72843.1 PCG72842.1 AGBW02014560 OWR41425.1 ODYU01011319 SOQ56990.1 APGK01042241 APGK01042242 APGK01042243 APGK01042244 APGK01042245 KB741002 ENN75782.1 KQ459583 KPI98674.1 JTDY01000301 KOB77804.1 GFDF01008376 JAV05708.1 CH964272 EDW84677.1 GANO01003491 JAB56380.1 CH933806 EDW16315.1 KQ415041 KOC58878.1 KRG02048.1 KRG02049.1 KRG00095.1 OUUW01000005 SPP80278.1 CH916373 EDV94410.1 CH940650 EDW67320.1 KRG02047.1 CH479179 EDW24209.1 GDHF01019078 JAI33236.1 GEBQ01000659 JAT39318.1 GAKP01013808 JAC45144.1 GDHF01032216 GDHF01029019 GDHF01028658 GDHF01023321 JAI20098.1 JAI23295.1 JAI23656.1 JAI28993.1 GDHF01023326 JAI28988.1 CH954182 EDV54197.1 CH902617 KPU79807.1 KRG02046.1 KRF83242.1 CM000070 EAL28114.3 KRT00466.1 KPU79805.1 EDV42586.1 KRT00468.1 KPU79806.1 KRF83241.1 KRF83244.1 KRF83245.1 KPU79802.1 KPU79804.1 KPU79803.1 KRT00467.1 KRT00469.1 CVRI01000070 CRL07100.1 APCN01003407 AXCM01004588 GDHC01008753 JAQ09876.1 GGFK01007987 MBW41308.1 AXCN02000736 AAAB01008984 ATLV01025637 KE525396 KFB52500.1
KPJ19460.1 GAIX01000626 JAA91934.1 PCG72839.1 PCG72843.1 PCG72842.1 AGBW02014560 OWR41425.1 ODYU01011319 SOQ56990.1 APGK01042241 APGK01042242 APGK01042243 APGK01042244 APGK01042245 KB741002 ENN75782.1 KQ459583 KPI98674.1 JTDY01000301 KOB77804.1 GFDF01008376 JAV05708.1 CH964272 EDW84677.1 GANO01003491 JAB56380.1 CH933806 EDW16315.1 KQ415041 KOC58878.1 KRG02048.1 KRG02049.1 KRG00095.1 OUUW01000005 SPP80278.1 CH916373 EDV94410.1 CH940650 EDW67320.1 KRG02047.1 CH479179 EDW24209.1 GDHF01019078 JAI33236.1 GEBQ01000659 JAT39318.1 GAKP01013808 JAC45144.1 GDHF01032216 GDHF01029019 GDHF01028658 GDHF01023321 JAI20098.1 JAI23295.1 JAI23656.1 JAI28993.1 GDHF01023326 JAI28988.1 CH954182 EDV54197.1 CH902617 KPU79807.1 KRG02046.1 KRF83242.1 CM000070 EAL28114.3 KRT00466.1 KPU79805.1 EDV42586.1 KRT00468.1 KPU79806.1 KRF83241.1 KRF83244.1 KRF83245.1 KPU79802.1 KPU79804.1 KPU79803.1 KRT00467.1 KRT00469.1 CVRI01000070 CRL07100.1 APCN01003407 AXCM01004588 GDHC01008753 JAQ09876.1 GGFK01007987 MBW41308.1 AXCN02000736 AAAB01008984 ATLV01025637 KE525396 KFB52500.1
Proteomes
UP000218220
UP000053240
UP000007151
UP000192223
UP000019118
UP000053268
+ More
UP000037510 UP000007798 UP000009192 UP000053825 UP000268350 UP000001070 UP000008792 UP000008744 UP000008711 UP000007801 UP000001819 UP000075885 UP000183832 UP000075902 UP000075840 UP000075883 UP000075903 UP000076407 UP000075900 UP000075882 UP000075920 UP000075886 UP000030765
UP000037510 UP000007798 UP000009192 UP000053825 UP000268350 UP000001070 UP000008792 UP000008744 UP000008711 UP000007801 UP000001819 UP000075885 UP000183832 UP000075902 UP000075840 UP000075883 UP000075903 UP000076407 UP000075900 UP000075882 UP000075920 UP000075886 UP000030765
Interpro
SUPFAM
SSF54791
SSF54791
Gene 3D
ProteinModelPortal
A0A2W1C369
A0A2A4JN60
A0A2A4JLI9
A0A194RQ63
S4PLE9
A0A2A4JLZ6
+ More
A0A2A4JN30 A0A2A4JN54 A0A212EIX1 A0A2H1WV98 A0A1W4WTR5 N6UAX0 A0A194Q109 A0A0L7LQX0 A0A1W4WTR2 A0A1L8DGX2 B4NHR7 U5ERZ3 B4K6L5 A0A0L7QJT4 A0A0Q9XCC9 A0A0Q9WVE0 A0A3B0K9Q1 B4JRM0 B4M0M8 A0A0Q9X1V2 B4G314 A0A0K8V329 A0A1B6MTS9 A0A034VRQ6 A0A0K8U0M7 A0A0K8UR47 B3P7N6 A0A0P8ZYV7 A0A0Q9XBX3 A0A0Q9WFQ1 Q297U1 A0A0R3NHA9 A0A0P8YLR8 B3LVL6 A0A0R3NHJ4 A0A0P9C8K6 A0A0Q9WF42 A0A0P8XYV5 A0A0P8XYF7 A0A0R3NN08 A0A182PVU3 A0A1J1J5M5 A0A182U5H2 A0A182I7I4 A0A182LS51 A0A182VNH4 A0A182WXM0 A0A182RZB5 A0A182LMK0 A0A182WGB8 A0A146LT23 A0A2M4AL17 A0A182QCU5 A0A1S4GZS9 A0A084WQK6
A0A2A4JN30 A0A2A4JN54 A0A212EIX1 A0A2H1WV98 A0A1W4WTR5 N6UAX0 A0A194Q109 A0A0L7LQX0 A0A1W4WTR2 A0A1L8DGX2 B4NHR7 U5ERZ3 B4K6L5 A0A0L7QJT4 A0A0Q9XCC9 A0A0Q9WVE0 A0A3B0K9Q1 B4JRM0 B4M0M8 A0A0Q9X1V2 B4G314 A0A0K8V329 A0A1B6MTS9 A0A034VRQ6 A0A0K8U0M7 A0A0K8UR47 B3P7N6 A0A0P8ZYV7 A0A0Q9XBX3 A0A0Q9WFQ1 Q297U1 A0A0R3NHA9 A0A0P8YLR8 B3LVL6 A0A0R3NHJ4 A0A0P9C8K6 A0A0Q9WF42 A0A0P8XYV5 A0A0P8XYF7 A0A0R3NN08 A0A182PVU3 A0A1J1J5M5 A0A182U5H2 A0A182I7I4 A0A182LS51 A0A182VNH4 A0A182WXM0 A0A182RZB5 A0A182LMK0 A0A182WGB8 A0A146LT23 A0A2M4AL17 A0A182QCU5 A0A1S4GZS9 A0A084WQK6
PDB
4JVH
E-value=3.31162e-54,
Score=533
Ontologies
GO
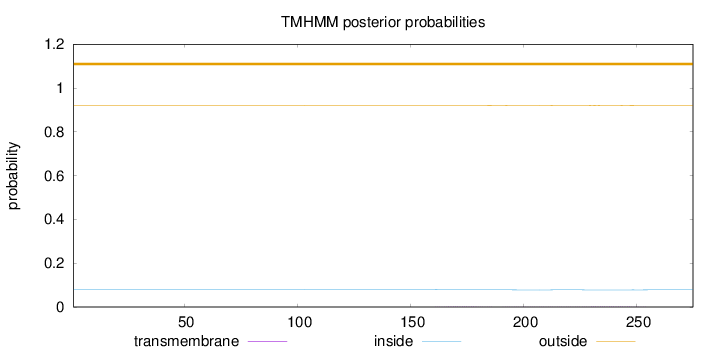
Topology
Length:
275
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0902400000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.07982
outside
1 - 275
Population Genetic Test Statistics
Pi
223.430025
Theta
185.310513
Tajima's D
0.445936
CLR
0.592391
CSRT
0.495825208739563
Interpretation
Uncertain
