Gene
KWMTBOMO03485
Annotation
PREDICTED:_non-structural_maintenance_of_chromosomes_element_1_homolog_[Amyelois_transitella]
Full name
4-hydroxybenzoate polyprenyltransferase, mitochondrial
+ More
Non-structural maintenance of chromosomes element 1 homolog
Non-structural maintenance of chromosomes element 1 homolog
Alternative Name
Para-hydroxybenzoate--polyprenyltransferase
Location in the cell
Nuclear Reliability : 3.506
Sequence
CDS
ATGTCTTACAGTAAAGTTCACAGATATTTTTTAAGAACTATAGCCAGTAGCGGTGTTATGAGTGTTGATGAAGCTCATCGAGTCCTGACTAAATTTACAGGTGAGCCGATCTTGTTAAAGAACTGCATTCAAGAAATTAATGATGTTATCAGACCATATCAGCAGTCAATCAAAGTAACATACAATGAAAGTACAAATGAAGAAGTAGTTATATTCTTAACCCATGCTTGGGATTCAATATCTGAATCCCAAAAGATCTTCACAGCTAAGGAATTAGACTATTTCAGATTATTAATTGAAGAAATAATGAGTATAGATGTAAGAGAAATCACAGGCATTCATGCAATTAATTTGGTTAATAAAATGGGCCAGAAAATATCAATGCTCCAAGCTGAAAATTTATTGGCTAAATGGTGTAAAATGTACTACTTAGAGAAGATACAAAATAAATACCGCTTAGGTGTGAGAACCATAAATGAATTTGAAGATTATTTTGAGAAATACATGCCTGATGTAATGAAAGATTGTTACATTTGTAAGAAAAATGTTTTTTGGGGGGCTAACTGCATAGCATGTGAAAAAGCTATTCACACTAGGTGCCTTAATCAGTACTTAGAAAAAATAAAAAAATGGCCTTGTTGTAAACTCGACTTCAGTCCAGATCAACTGAAACAACTTAACGAAAGCCAGAGAATGACTCAGATAACTCAAACCTTTGAAGACACACAGTCAATTGAAGAAAGGTCGGAGAATTCAACAGCTTCCAATGTAGAAAATGATAATACAGAAGAAATGGAAAGAAGTCAAAATACAGATAACATACGACGTCTCTCTAGAAAGAGGAAGAGAGAATAA
Protein
MSYSKVHRYFLRTIASSGVMSVDEAHRVLTKFTGEPILLKNCIQEINDVIRPYQQSIKVTYNESTNEEVVIFLTHAWDSISESQKIFTAKELDYFRLLIEEIMSIDVREITGIHAINLVNKMGQKISMLQAENLLAKWCKMYYLEKIQNKYRLGVRTINEFEDYFEKYMPDVMKDCYICKKNVFWGANCIACEKAIHTRCLNQYLEKIKKWPCCKLDFSPDQLKQLNESQRMTQITQTFEDTQSIEERSENSTASNVENDNTEEMERSQNTDNIRRLSRKRKRE
Summary
Description
Catalyzes the prenylation of para-hydroxybenzoate (PHB) with an all-trans polyprenyl group. Mediates the second step in the final reaction sequence of coenzyme Q (CoQ) biosynthesis, which is the condensation of the polyisoprenoid side chain with PHB, generating the first membrane-bound Q intermediate.
RING-type zinc finger-containing E3 ubiquitin ligase that assembles with melanoma antigen protein (MAGE) to catalyze the direct transfer of ubiquitin from E2 ubiquitin-conjugating enzyme to a specific substrate. Within MAGE-RING ubiquitin ligase complex, MAGE stimulates and specifies ubiquitin ligase activity likely through recruitment and/or stabilization of the E2 ubiquitin-conjugating enzyme at the E3:substrate complex. Involved in maintenance of genome integrity, DNA damage response and DNA repair. NSMCE3/MAGEG1 and NSMCE1 ubiquitin ligase are components of SMC5-SMC6 complex and may positively regulate homologous recombination-mediated DNA repair.
RING-type zinc finger-containing E3 ubiquitin ligase that assembles with melanoma antigen protein (MAGE) to catalyze the direct transfer of ubiquitin from E2 ubiquitin-conjugating enzyme to a specific substrate. Within MAGE-RING ubiquitin ligase complex, MAGE stimulates and specifies ubiquitin ligase activity likely through recruitment and/or stabilization of the E2 ubiquitin-conjugating enzyme at the E3:substrate complex. Involved in maintenance of genome integrity, DNA damage response and DNA repair. NSMCE3/MAGEG1 and NSMCE1 ubiquitin ligase are components of SMC5-SMC6 complex and may positively regulate homologous recombination-mediated DNA repair.
Catalytic Activity
4-hydroxybenzoate + an all-trans-polyprenyl diphosphate = a 4-hydroxy-3-all-trans-polyprenylbenzoate + diphosphate
S-ubiquitinyl-[E2 ubiquitin-conjugating enzyme]-L-cysteine + [acceptor protein]-L-lysine = [E2 ubiquitin-conjugating enzyme]-L-cysteine + N(6)-ubiquitinyl-[acceptor protein]-L-lysine.
S-ubiquitinyl-[E2 ubiquitin-conjugating enzyme]-L-cysteine + [acceptor protein]-L-lysine = [E2 ubiquitin-conjugating enzyme]-L-cysteine + N(6)-ubiquitinyl-[acceptor protein]-L-lysine.
Cofactor
Mg(2+)
Subunit
Component of the SMC5-SMC6 complex which consists at least of SMC5, SMC6, NSMCE2, NSMCE1, NSMCE4A or EID3 and NSMCE3. NSMCE1, NSMCE4A or EID3 and NSMCE3 probably form a subcomplex that bridges the head domains of the SMC5-SMC6 heterodimer. Interacts with NSMCE3.
Similarity
Belongs to the calycin superfamily. Lipocalin family.
Belongs to the CDP-alcohol phosphatidyltransferase class-I family.
Belongs to the UbiA prenyltransferase family.
Belongs to the NSE1 family.
Belongs to the CDP-alcohol phosphatidyltransferase class-I family.
Belongs to the UbiA prenyltransferase family.
Belongs to the NSE1 family.
Keywords
Chromosome
Complete proteome
DNA damage
DNA recombination
DNA repair
Metal-binding
Nucleus
Reference proteome
Telomere
Transferase
Ubl conjugation
Ubl conjugation pathway
Zinc
Zinc-finger
Feature
chain Non-structural maintenance of chromosomes element 1 homolog
Uniprot
H9JAI5
A0A2W1BW85
A0A2A4JL89
A0A0L7LQM6
A0A2H1WNW9
A0A194PZC2
+ More
A0A194RPM2 A0A1E1WP66 S4PRT9 A0A212EWM9 A0A084W2U7 A0A182NM98 A0A182IY62 W5JVK0 Q0IFZ1 A0A182FFD2 A0A1Y1MUU0 A0A023EMY4 A0A182GQK8 A0A0K8TLT2 A0A182V699 A0A182MTZ3 Q16PA4 A0A182QE16 B0WPT4 Q7PNL4 A0A182LJP5 A0A182HM54 A0A182Y0U5 A0A182PN97 A0A1S4FU51 A0A182WK80 A0A182RQ96 A0A182XCS8 A0A1Q3F251 A0A182K710 A0A139WF15 A0A182GPD4 A0A1B0D601 B0X5P6 A0A1J1I5W1 A0A182UIE1 A0A1W4WZ49 N6TT82 A0A1I8NHF5 J4G7D8 A0A3L8DA19 A0A226E201 A0A1D2MQ27 A0A195E5H4 K7IS45 A0A1I8PFM2 A0A232EW33 A0A384BLK0 A0A384BLI6 A0A369JNE1 A0A341BAW9 R7V2F2 B4KJU9 E2ADT0 A0A195B0G5 D2I208 A0A158NQZ2 A0A1S3ERR1 A0A151WMC6 A0A2Y9PN56 A0A195EVG1 G1LBK8 B4JPS2 E2RFI7 A0A0L7QQF7 I3MYE7 G1DFX6 B3MKT7 A0A2Y9PLL6 H0WQR7 A0A2U3VPV4 M3YNH9 A0A2Y9L8G0 Q3T0X7 A0A2Y9LKS2 A0A2Y9LAS1 A0A2U3ZCR9 A0A3Q0CVU7 A0A2K6BS10 L5KNB2 A0A2Y9HX60 G7NQF5 M3W893 G7Q0S6 B2RDU2 A0A1U7UIK6 A0A2K6GY88 A0A2A3ECE5 A0A340WYY9 A0A2K6P830 A0A2K5VW03 H9FQY0
A0A194RPM2 A0A1E1WP66 S4PRT9 A0A212EWM9 A0A084W2U7 A0A182NM98 A0A182IY62 W5JVK0 Q0IFZ1 A0A182FFD2 A0A1Y1MUU0 A0A023EMY4 A0A182GQK8 A0A0K8TLT2 A0A182V699 A0A182MTZ3 Q16PA4 A0A182QE16 B0WPT4 Q7PNL4 A0A182LJP5 A0A182HM54 A0A182Y0U5 A0A182PN97 A0A1S4FU51 A0A182WK80 A0A182RQ96 A0A182XCS8 A0A1Q3F251 A0A182K710 A0A139WF15 A0A182GPD4 A0A1B0D601 B0X5P6 A0A1J1I5W1 A0A182UIE1 A0A1W4WZ49 N6TT82 A0A1I8NHF5 J4G7D8 A0A3L8DA19 A0A226E201 A0A1D2MQ27 A0A195E5H4 K7IS45 A0A1I8PFM2 A0A232EW33 A0A384BLK0 A0A384BLI6 A0A369JNE1 A0A341BAW9 R7V2F2 B4KJU9 E2ADT0 A0A195B0G5 D2I208 A0A158NQZ2 A0A1S3ERR1 A0A151WMC6 A0A2Y9PN56 A0A195EVG1 G1LBK8 B4JPS2 E2RFI7 A0A0L7QQF7 I3MYE7 G1DFX6 B3MKT7 A0A2Y9PLL6 H0WQR7 A0A2U3VPV4 M3YNH9 A0A2Y9L8G0 Q3T0X7 A0A2Y9LKS2 A0A2Y9LAS1 A0A2U3ZCR9 A0A3Q0CVU7 A0A2K6BS10 L5KNB2 A0A2Y9HX60 G7NQF5 M3W893 G7Q0S6 B2RDU2 A0A1U7UIK6 A0A2K6GY88 A0A2A3ECE5 A0A340WYY9 A0A2K6P830 A0A2K5VW03 H9FQY0
EC Number
2.5.1.39
2.3.2.27
2.3.2.27
Pubmed
19121390
28756777
26227816
26354079
23622113
22118469
+ More
24438588 20920257 23761445 17510324 28004739 24945155 26483478 26369729 12364791 14747013 17210077 20966253 25244985 18362917 19820115 23537049 25315136 22247176 30249741 27289101 20075255 28648823 24813606 23254933 17994087 20798317 20010809 21347285 16341006 26319212 23258410 22002653 17975172 25362486 25319552
24438588 20920257 23761445 17510324 28004739 24945155 26483478 26369729 12364791 14747013 17210077 20966253 25244985 18362917 19820115 23537049 25315136 22247176 30249741 27289101 20075255 28648823 24813606 23254933 17994087 20798317 20010809 21347285 16341006 26319212 23258410 22002653 17975172 25362486 25319552
EMBL
BABH01009435
BABH01009436
BABH01009437
KZ149899
PZC78601.1
NWSH01001063
+ More
PCG72845.1 JTDY01000301 KOB77803.1 ODYU01009962 SOQ54728.1 KQ459583 KPI98671.1 KQ459896 KPJ19457.1 GDQN01002423 JAT88631.1 GAIX01000520 JAA92040.1 AGBW02011942 OWR45879.1 ATLV01019698 KE525278 KFB44541.1 ADMH02000355 ETN66854.1 CH477281 EAT44933.1 GEZM01020066 JAV89421.1 GAPW01003884 JAC09714.1 JXUM01015651 KQ560436 KXJ82427.1 GDAI01002271 JAI15332.1 AXCM01001822 CH477789 EAT36198.1 AXCN02000938 DS232029 EDS32499.1 AAAB01008960 EAA11970.5 APCN01004355 GFDL01013467 JAV21578.1 KQ971354 KYB26512.1 JXUM01078212 KQ563051 KXJ74565.1 AJVK01011901 AJVK01011902 DS232391 EDS40983.1 CVRI01000042 CRK95571.1 APGK01019841 APGK01053910 KB741239 KB740134 KB632275 ENN72114.1 ENN81278.1 ERL91088.1 HE797078 CCM02353.1 QOIP01000010 RLU17327.1 LNIX01000008 OXA51310.1 LJIJ01000697 ODM95209.1 KQ979608 KYN20440.1 NNAY01001922 OXU22562.1 LUEZ02000055 RDB21203.1 AMQN01005336 KB295744 ELU12684.1 CH933807 EDW12552.1 GL438820 EFN68442.1 KQ976691 KYM77951.1 GL194078 EFB28138.1 ADTU01023710 KQ982944 KYQ48980.1 KQ981965 KYN31874.1 ACTA01017866 CH916372 EDV98902.1 AAEX03004424 KQ414786 KOC60809.1 AGTP01031047 AGTP01031048 JI861692 AEJ84180.1 CH902620 EDV31618.1 AAQR03122989 AAQR03122990 AEYP01082357 AEYP01082358 AEYP01082359 BC102214 KB030664 ELK12078.1 CM001272 EHH31533.1 AANG04002788 CM001295 EHH60264.1 AK315674 BAG38039.1 KZ288295 PBC28962.1 AQIA01042469 JU333284 JU474598 JV046378 AFE77039.1 AFH31402.1 AFI36449.1
PCG72845.1 JTDY01000301 KOB77803.1 ODYU01009962 SOQ54728.1 KQ459583 KPI98671.1 KQ459896 KPJ19457.1 GDQN01002423 JAT88631.1 GAIX01000520 JAA92040.1 AGBW02011942 OWR45879.1 ATLV01019698 KE525278 KFB44541.1 ADMH02000355 ETN66854.1 CH477281 EAT44933.1 GEZM01020066 JAV89421.1 GAPW01003884 JAC09714.1 JXUM01015651 KQ560436 KXJ82427.1 GDAI01002271 JAI15332.1 AXCM01001822 CH477789 EAT36198.1 AXCN02000938 DS232029 EDS32499.1 AAAB01008960 EAA11970.5 APCN01004355 GFDL01013467 JAV21578.1 KQ971354 KYB26512.1 JXUM01078212 KQ563051 KXJ74565.1 AJVK01011901 AJVK01011902 DS232391 EDS40983.1 CVRI01000042 CRK95571.1 APGK01019841 APGK01053910 KB741239 KB740134 KB632275 ENN72114.1 ENN81278.1 ERL91088.1 HE797078 CCM02353.1 QOIP01000010 RLU17327.1 LNIX01000008 OXA51310.1 LJIJ01000697 ODM95209.1 KQ979608 KYN20440.1 NNAY01001922 OXU22562.1 LUEZ02000055 RDB21203.1 AMQN01005336 KB295744 ELU12684.1 CH933807 EDW12552.1 GL438820 EFN68442.1 KQ976691 KYM77951.1 GL194078 EFB28138.1 ADTU01023710 KQ982944 KYQ48980.1 KQ981965 KYN31874.1 ACTA01017866 CH916372 EDV98902.1 AAEX03004424 KQ414786 KOC60809.1 AGTP01031047 AGTP01031048 JI861692 AEJ84180.1 CH902620 EDV31618.1 AAQR03122989 AAQR03122990 AEYP01082357 AEYP01082358 AEYP01082359 BC102214 KB030664 ELK12078.1 CM001272 EHH31533.1 AANG04002788 CM001295 EHH60264.1 AK315674 BAG38039.1 KZ288295 PBC28962.1 AQIA01042469 JU333284 JU474598 JV046378 AFE77039.1 AFH31402.1 AFI36449.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053268
UP000053240
UP000007151
+ More
UP000030765 UP000075884 UP000075880 UP000000673 UP000008820 UP000069272 UP000069940 UP000249989 UP000075903 UP000075883 UP000075886 UP000002320 UP000007062 UP000075882 UP000075840 UP000076408 UP000075885 UP000075920 UP000075900 UP000076407 UP000075881 UP000007266 UP000092462 UP000183832 UP000075902 UP000192223 UP000019118 UP000030742 UP000095301 UP000006352 UP000279307 UP000198287 UP000094527 UP000078492 UP000002358 UP000095300 UP000215335 UP000261680 UP000291021 UP000076154 UP000252040 UP000014760 UP000009192 UP000000311 UP000078540 UP000005205 UP000081671 UP000075809 UP000248483 UP000078541 UP000008912 UP000001070 UP000002254 UP000053825 UP000005215 UP000007801 UP000005225 UP000245340 UP000000715 UP000248482 UP000009136 UP000189706 UP000233120 UP000010552 UP000248481 UP000011712 UP000009130 UP000189704 UP000233160 UP000242457 UP000265300 UP000233200 UP000233100
UP000030765 UP000075884 UP000075880 UP000000673 UP000008820 UP000069272 UP000069940 UP000249989 UP000075903 UP000075883 UP000075886 UP000002320 UP000007062 UP000075882 UP000075840 UP000076408 UP000075885 UP000075920 UP000075900 UP000076407 UP000075881 UP000007266 UP000092462 UP000183832 UP000075902 UP000192223 UP000019118 UP000030742 UP000095301 UP000006352 UP000279307 UP000198287 UP000094527 UP000078492 UP000002358 UP000095300 UP000215335 UP000261680 UP000291021 UP000076154 UP000252040 UP000014760 UP000009192 UP000000311 UP000078540 UP000005205 UP000081671 UP000075809 UP000248483 UP000078541 UP000008912 UP000001070 UP000002254 UP000053825 UP000005215 UP000007801 UP000005225 UP000245340 UP000000715 UP000248482 UP000009136 UP000189706 UP000233120 UP000010552 UP000248481 UP000011712 UP000009130 UP000189704 UP000233160 UP000242457 UP000265300 UP000233200 UP000233100
Pfam
Interpro
IPR011513
Nse1
+ More
IPR036388 WH-like_DNA-bd_sf
IPR002219 PE/DAG-bd
IPR013083 Znf_RING/FYVE/PHD
IPR014857 Znf_RING-like
IPR001841 Znf_RING
IPR019786 Zinc_finger_PHD-type_CS
IPR022272 Lipocalin_CS
IPR000566 Lipocln_cytosolic_FA-bd_dom
IPR012674 Calycin
IPR014472 CHOPT
IPR000462 CDP-OH_P_trans
IPR006370 HB_polyprenyltransferase-like
IPR000537 UbiA_prenyltransferase
IPR030470 UbiA_prenylTrfase_CS
IPR011011 Znf_FYVE_PHD
IPR036388 WH-like_DNA-bd_sf
IPR002219 PE/DAG-bd
IPR013083 Znf_RING/FYVE/PHD
IPR014857 Znf_RING-like
IPR001841 Znf_RING
IPR019786 Zinc_finger_PHD-type_CS
IPR022272 Lipocalin_CS
IPR000566 Lipocln_cytosolic_FA-bd_dom
IPR012674 Calycin
IPR014472 CHOPT
IPR000462 CDP-OH_P_trans
IPR006370 HB_polyprenyltransferase-like
IPR000537 UbiA_prenyltransferase
IPR030470 UbiA_prenylTrfase_CS
IPR011011 Znf_FYVE_PHD
Gene 3D
ProteinModelPortal
H9JAI5
A0A2W1BW85
A0A2A4JL89
A0A0L7LQM6
A0A2H1WNW9
A0A194PZC2
+ More
A0A194RPM2 A0A1E1WP66 S4PRT9 A0A212EWM9 A0A084W2U7 A0A182NM98 A0A182IY62 W5JVK0 Q0IFZ1 A0A182FFD2 A0A1Y1MUU0 A0A023EMY4 A0A182GQK8 A0A0K8TLT2 A0A182V699 A0A182MTZ3 Q16PA4 A0A182QE16 B0WPT4 Q7PNL4 A0A182LJP5 A0A182HM54 A0A182Y0U5 A0A182PN97 A0A1S4FU51 A0A182WK80 A0A182RQ96 A0A182XCS8 A0A1Q3F251 A0A182K710 A0A139WF15 A0A182GPD4 A0A1B0D601 B0X5P6 A0A1J1I5W1 A0A182UIE1 A0A1W4WZ49 N6TT82 A0A1I8NHF5 J4G7D8 A0A3L8DA19 A0A226E201 A0A1D2MQ27 A0A195E5H4 K7IS45 A0A1I8PFM2 A0A232EW33 A0A384BLK0 A0A384BLI6 A0A369JNE1 A0A341BAW9 R7V2F2 B4KJU9 E2ADT0 A0A195B0G5 D2I208 A0A158NQZ2 A0A1S3ERR1 A0A151WMC6 A0A2Y9PN56 A0A195EVG1 G1LBK8 B4JPS2 E2RFI7 A0A0L7QQF7 I3MYE7 G1DFX6 B3MKT7 A0A2Y9PLL6 H0WQR7 A0A2U3VPV4 M3YNH9 A0A2Y9L8G0 Q3T0X7 A0A2Y9LKS2 A0A2Y9LAS1 A0A2U3ZCR9 A0A3Q0CVU7 A0A2K6BS10 L5KNB2 A0A2Y9HX60 G7NQF5 M3W893 G7Q0S6 B2RDU2 A0A1U7UIK6 A0A2K6GY88 A0A2A3ECE5 A0A340WYY9 A0A2K6P830 A0A2K5VW03 H9FQY0
A0A194RPM2 A0A1E1WP66 S4PRT9 A0A212EWM9 A0A084W2U7 A0A182NM98 A0A182IY62 W5JVK0 Q0IFZ1 A0A182FFD2 A0A1Y1MUU0 A0A023EMY4 A0A182GQK8 A0A0K8TLT2 A0A182V699 A0A182MTZ3 Q16PA4 A0A182QE16 B0WPT4 Q7PNL4 A0A182LJP5 A0A182HM54 A0A182Y0U5 A0A182PN97 A0A1S4FU51 A0A182WK80 A0A182RQ96 A0A182XCS8 A0A1Q3F251 A0A182K710 A0A139WF15 A0A182GPD4 A0A1B0D601 B0X5P6 A0A1J1I5W1 A0A182UIE1 A0A1W4WZ49 N6TT82 A0A1I8NHF5 J4G7D8 A0A3L8DA19 A0A226E201 A0A1D2MQ27 A0A195E5H4 K7IS45 A0A1I8PFM2 A0A232EW33 A0A384BLK0 A0A384BLI6 A0A369JNE1 A0A341BAW9 R7V2F2 B4KJU9 E2ADT0 A0A195B0G5 D2I208 A0A158NQZ2 A0A1S3ERR1 A0A151WMC6 A0A2Y9PN56 A0A195EVG1 G1LBK8 B4JPS2 E2RFI7 A0A0L7QQF7 I3MYE7 G1DFX6 B3MKT7 A0A2Y9PLL6 H0WQR7 A0A2U3VPV4 M3YNH9 A0A2Y9L8G0 Q3T0X7 A0A2Y9LKS2 A0A2Y9LAS1 A0A2U3ZCR9 A0A3Q0CVU7 A0A2K6BS10 L5KNB2 A0A2Y9HX60 G7NQF5 M3W893 G7Q0S6 B2RDU2 A0A1U7UIK6 A0A2K6GY88 A0A2A3ECE5 A0A340WYY9 A0A2K6P830 A0A2K5VW03 H9FQY0
PDB
5WY5
E-value=3.19343e-14,
Score=189
Ontologies
GO
Topology
Subcellular location
Mitochondrion inner membrane
Nucleus
Chromosome
Telomere
Nucleus
Chromosome
Telomere
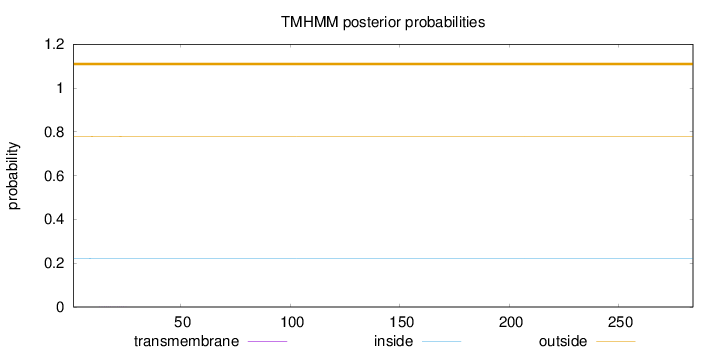
Length:
284
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00563999999999999
Exp number, first 60 AAs:
0.00263
Total prob of N-in:
0.22188
outside
1 - 284
Population Genetic Test Statistics
Pi
323.025637
Theta
200.746511
Tajima's D
0.610292
CLR
0.00042
CSRT
0.548422578871056
Interpretation
Uncertain
