Gene
KWMTBOMO03478
Pre Gene Modal
BGIBMGA006528
Annotation
PREDICTED:_lachesin-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.057
Sequence
CDS
ATGGGTCATCTTTCAGTGGTGATACCGCCAGAGATAGCCGATGAAGATGGATCGGAAGCAAGTGCTCCTGAAGGTGGGTCGGTGGAACTGCGGTGTACAGCTTCCGGGGTGCCTGAACCTAACATATCTTGGAAGCGGACCTTGGGTAGGAACATTATCCTGCGTGACAGTAATGGACAGGAACTGAAAGTGGTGGACAGCTATGTTGGTTCAACTCTTGTGCTGAAAGGCCTGCGGCGTTCGGACATGGGCACCTATCTGTGTATCGCAGCCAACGGCATTCCGCCTACAAAGAGCCGCCGCTACGAGGTCACCGTACTCTGTGAGTAA
Protein
MGHLSVVIPPEIADEDGSEASAPEGGSVELRCTASGVPEPNISWKRTLGRNIILRDSNGQELKVVDSYVGSTLVLKGLRRSDMGTYLCIAANGIPPTKSRRYEVTVLCE
Summary
Uniprot
H9JAI3
A0A2H1VT25
A0A2W1BLT9
A0A194RQ57
A0A194Q104
A0A212F663
+ More
A0A2A4K0S7 A0A2W1BJM5 A0A2A4K0W3 A0A2A4JZ35 A0A194Q5K0 A0A212F656 A0A194RTL3 K7J3Y4 A0A1Y1LUS4 A0A0N0BBD3 A0A088AJG9 E2AYG8 A0A195F292 A0A3L8DWQ2 E2BDH1 F4X3S8 A0A026WPA9 A0A195DZY3 A0A067R5S2 A0A151XJQ1 A0A195BGK8 A0A195CRF6 A0A2A3E6U9 A0A088ALP5 A0A1J1IRA3 E2ACI2 A0A0L7QPA4 A0A182SH50 N6TI71 A0A0M9A176 A0A139WP45 A0A182YRZ5 A0A1B0GM78 A0A310SMD6 A0A194PUH5 A0A336LZ60 A0A182WFE3 A0A151J7V8 A0A154P8T4 A0A151I737 A0A182NGR2 K7IZ51 A0A151WP42 A0A182QJ96 A0A182UPK0 A0A1S4GAQ9 A0A212FI49 E2BBN1 A0A3Q0JKY0 A0A195BVQ0 A0A182IBH9 A0A182X5U6 A0A182RK29 A0NFK2 A0A026W6I4 A0A084VSN0 A0A182MPH9 A0A3L8DSL2 A0A182TKB1 E0VQ13 A0A158N921 A0A2H1WU64 A0A2S2Q2I9 A0A154PIL3 A0A182W618 A0A2A4K4N0 A0A0L7QRK4 A0A2H8TMT1 A0A182RVN5 A0A0L0BQE5 A0A1B0CRC5 A0A195FUG6 A0A1B0FJ66 A0A1A9Z437 B4LQZ3 A0A1B0BQH0 A0A2H1W1K5 A0A182PTH7 A0A0L0C9B6 F4WHI3 A0A2M4CNM6 A0A2A4JL61 A0A182J2A1 A0A1I8MHH7 A0A0J7L7S8 A0A2M4BMW1 A0A1B0FFM0 A0A2A4JJQ6 A0A1I8NHV3 A0A3B0JU01 A0A0Q9X6E4 A0A3B0JWZ3 A0A1I8NN68
A0A2A4K0S7 A0A2W1BJM5 A0A2A4K0W3 A0A2A4JZ35 A0A194Q5K0 A0A212F656 A0A194RTL3 K7J3Y4 A0A1Y1LUS4 A0A0N0BBD3 A0A088AJG9 E2AYG8 A0A195F292 A0A3L8DWQ2 E2BDH1 F4X3S8 A0A026WPA9 A0A195DZY3 A0A067R5S2 A0A151XJQ1 A0A195BGK8 A0A195CRF6 A0A2A3E6U9 A0A088ALP5 A0A1J1IRA3 E2ACI2 A0A0L7QPA4 A0A182SH50 N6TI71 A0A0M9A176 A0A139WP45 A0A182YRZ5 A0A1B0GM78 A0A310SMD6 A0A194PUH5 A0A336LZ60 A0A182WFE3 A0A151J7V8 A0A154P8T4 A0A151I737 A0A182NGR2 K7IZ51 A0A151WP42 A0A182QJ96 A0A182UPK0 A0A1S4GAQ9 A0A212FI49 E2BBN1 A0A3Q0JKY0 A0A195BVQ0 A0A182IBH9 A0A182X5U6 A0A182RK29 A0NFK2 A0A026W6I4 A0A084VSN0 A0A182MPH9 A0A3L8DSL2 A0A182TKB1 E0VQ13 A0A158N921 A0A2H1WU64 A0A2S2Q2I9 A0A154PIL3 A0A182W618 A0A2A4K4N0 A0A0L7QRK4 A0A2H8TMT1 A0A182RVN5 A0A0L0BQE5 A0A1B0CRC5 A0A195FUG6 A0A1B0FJ66 A0A1A9Z437 B4LQZ3 A0A1B0BQH0 A0A2H1W1K5 A0A182PTH7 A0A0L0C9B6 F4WHI3 A0A2M4CNM6 A0A2A4JL61 A0A182J2A1 A0A1I8MHH7 A0A0J7L7S8 A0A2M4BMW1 A0A1B0FFM0 A0A2A4JJQ6 A0A1I8NHV3 A0A3B0JU01 A0A0Q9X6E4 A0A3B0JWZ3 A0A1I8NN68
Pubmed
EMBL
BABH01009416
BABH01009417
ODYU01003976
SOQ43384.1
KZ150004
PZC75251.1
+ More
KQ459896 KPJ19455.1 KQ459583 KPI98669.1 AGBW02010073 OWR49223.1 NWSH01000340 PCG77280.1 PZC75249.1 PCG77282.1 PCG77281.1 KPI98670.1 OWR49222.1 KPJ19456.1 AAZX01004985 GEZM01051704 JAV74737.1 KQ436131 KOX67431.1 GL443910 EFN61503.1 KQ981856 KYN34501.1 QOIP01000003 RLU24663.1 GL447620 EFN86283.1 GL888624 EGI58875.1 KK107139 EZA57855.1 KQ979955 KYN18470.1 KK852725 KDR17669.1 KQ982074 KYQ60508.1 KQ976490 KYM83356.1 KQ977394 KYN03077.1 KZ288349 PBC27420.1 CVRI01000055 CRL01622.1 GL438528 EFN68848.1 KQ414821 KOC60460.1 APGK01036801 KB740943 KB632399 ENN77518.1 ERL94858.1 KQ435794 KOX73793.1 KQ971307 KYB29768.1 AJVK01011203 AJVK01011204 AJVK01011205 KQ762190 OAD56151.1 KQ459597 KPI94790.1 UFQT01000326 SSX23274.1 KQ979640 KYN20030.1 KQ434827 KZC07628.1 KQ978451 KYM93933.1 KQ982907 KYQ49465.1 AXCN02002079 AAAB01008973 AGBW02008433 OWR53414.1 GL447128 EFN86908.1 KQ976405 KYM91883.1 APCN01000703 EAU76204.3 KK107372 EZA51722.1 ATLV01016092 ATLV01016093 ATLV01016094 KE525057 KFB40974.1 AXCM01008344 AXCM01008345 AXCM01008346 QOIP01000004 RLU23401.1 DS235389 EEB15469.1 ADTU01009044 ODYU01011088 SOQ56605.1 GGMS01002687 MBY71890.1 KQ434924 KZC11652.1 NWSH01000132 PCG79215.1 KQ414775 KOC61270.1 GFXV01003679 MBW15484.1 JRES01001533 KNC22208.1 AJWK01024595 AJWK01024596 AJWK01024597 KQ981268 KYN43937.1 CCAG010003433 CH940649 EDW64532.2 JXJN01018616 JXJN01018617 ODYU01005716 SOQ46836.1 JRES01000741 KNC28830.1 GL888161 EGI66337.1 GGFL01002746 MBW66924.1 NWSH01001156 PCG72324.1 AXCP01007149 LBMM01000345 KMR00004.1 GGFJ01005254 MBW54395.1 CCAG010008667 CCAG010008668 PCG72325.1 OUUW01000010 SPP85594.1 CH933807 KRG03819.1 SPP85593.1
KQ459896 KPJ19455.1 KQ459583 KPI98669.1 AGBW02010073 OWR49223.1 NWSH01000340 PCG77280.1 PZC75249.1 PCG77282.1 PCG77281.1 KPI98670.1 OWR49222.1 KPJ19456.1 AAZX01004985 GEZM01051704 JAV74737.1 KQ436131 KOX67431.1 GL443910 EFN61503.1 KQ981856 KYN34501.1 QOIP01000003 RLU24663.1 GL447620 EFN86283.1 GL888624 EGI58875.1 KK107139 EZA57855.1 KQ979955 KYN18470.1 KK852725 KDR17669.1 KQ982074 KYQ60508.1 KQ976490 KYM83356.1 KQ977394 KYN03077.1 KZ288349 PBC27420.1 CVRI01000055 CRL01622.1 GL438528 EFN68848.1 KQ414821 KOC60460.1 APGK01036801 KB740943 KB632399 ENN77518.1 ERL94858.1 KQ435794 KOX73793.1 KQ971307 KYB29768.1 AJVK01011203 AJVK01011204 AJVK01011205 KQ762190 OAD56151.1 KQ459597 KPI94790.1 UFQT01000326 SSX23274.1 KQ979640 KYN20030.1 KQ434827 KZC07628.1 KQ978451 KYM93933.1 KQ982907 KYQ49465.1 AXCN02002079 AAAB01008973 AGBW02008433 OWR53414.1 GL447128 EFN86908.1 KQ976405 KYM91883.1 APCN01000703 EAU76204.3 KK107372 EZA51722.1 ATLV01016092 ATLV01016093 ATLV01016094 KE525057 KFB40974.1 AXCM01008344 AXCM01008345 AXCM01008346 QOIP01000004 RLU23401.1 DS235389 EEB15469.1 ADTU01009044 ODYU01011088 SOQ56605.1 GGMS01002687 MBY71890.1 KQ434924 KZC11652.1 NWSH01000132 PCG79215.1 KQ414775 KOC61270.1 GFXV01003679 MBW15484.1 JRES01001533 KNC22208.1 AJWK01024595 AJWK01024596 AJWK01024597 KQ981268 KYN43937.1 CCAG010003433 CH940649 EDW64532.2 JXJN01018616 JXJN01018617 ODYU01005716 SOQ46836.1 JRES01000741 KNC28830.1 GL888161 EGI66337.1 GGFL01002746 MBW66924.1 NWSH01001156 PCG72324.1 AXCP01007149 LBMM01000345 KMR00004.1 GGFJ01005254 MBW54395.1 CCAG010008667 CCAG010008668 PCG72325.1 OUUW01000010 SPP85594.1 CH933807 KRG03819.1 SPP85593.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000218220
UP000002358
+ More
UP000053105 UP000005203 UP000000311 UP000078541 UP000279307 UP000008237 UP000007755 UP000053097 UP000078492 UP000027135 UP000075809 UP000078540 UP000078542 UP000242457 UP000183832 UP000053825 UP000075901 UP000019118 UP000030742 UP000007266 UP000076408 UP000092462 UP000075920 UP000076502 UP000075884 UP000075886 UP000075903 UP000079169 UP000075840 UP000076407 UP000075900 UP000007062 UP000030765 UP000075883 UP000075902 UP000009046 UP000005205 UP000037069 UP000092461 UP000092444 UP000092445 UP000008792 UP000092460 UP000075885 UP000075880 UP000095301 UP000036403 UP000268350 UP000009192 UP000095300
UP000053105 UP000005203 UP000000311 UP000078541 UP000279307 UP000008237 UP000007755 UP000053097 UP000078492 UP000027135 UP000075809 UP000078540 UP000078542 UP000242457 UP000183832 UP000053825 UP000075901 UP000019118 UP000030742 UP000007266 UP000076408 UP000092462 UP000075920 UP000076502 UP000075884 UP000075886 UP000075903 UP000079169 UP000075840 UP000076407 UP000075900 UP000007062 UP000030765 UP000075883 UP000075902 UP000009046 UP000005205 UP000037069 UP000092461 UP000092444 UP000092445 UP000008792 UP000092460 UP000075885 UP000075880 UP000095301 UP000036403 UP000268350 UP000009192 UP000095300
PRIDE
Pfam
Interpro
IPR036179
Ig-like_dom_sf
+ More
IPR003598 Ig_sub2
IPR013783 Ig-like_fold
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013098 Ig_I-set
IPR013106 Ig_V-set
IPR003006 Ig/MHC_CS
IPR032675 LRR_dom_sf
IPR003591 Leu-rich_rpt_typical-subtyp
IPR001611 Leu-rich_rpt
IPR013151 Immunoglobulin
IPR001148 CA_dom
IPR036398 CA_dom_sf
IPR041426 Mos1_HTH
IPR003598 Ig_sub2
IPR013783 Ig-like_fold
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013098 Ig_I-set
IPR013106 Ig_V-set
IPR003006 Ig/MHC_CS
IPR032675 LRR_dom_sf
IPR003591 Leu-rich_rpt_typical-subtyp
IPR001611 Leu-rich_rpt
IPR013151 Immunoglobulin
IPR001148 CA_dom
IPR036398 CA_dom_sf
IPR041426 Mos1_HTH
Gene 3D
ProteinModelPortal
H9JAI3
A0A2H1VT25
A0A2W1BLT9
A0A194RQ57
A0A194Q104
A0A212F663
+ More
A0A2A4K0S7 A0A2W1BJM5 A0A2A4K0W3 A0A2A4JZ35 A0A194Q5K0 A0A212F656 A0A194RTL3 K7J3Y4 A0A1Y1LUS4 A0A0N0BBD3 A0A088AJG9 E2AYG8 A0A195F292 A0A3L8DWQ2 E2BDH1 F4X3S8 A0A026WPA9 A0A195DZY3 A0A067R5S2 A0A151XJQ1 A0A195BGK8 A0A195CRF6 A0A2A3E6U9 A0A088ALP5 A0A1J1IRA3 E2ACI2 A0A0L7QPA4 A0A182SH50 N6TI71 A0A0M9A176 A0A139WP45 A0A182YRZ5 A0A1B0GM78 A0A310SMD6 A0A194PUH5 A0A336LZ60 A0A182WFE3 A0A151J7V8 A0A154P8T4 A0A151I737 A0A182NGR2 K7IZ51 A0A151WP42 A0A182QJ96 A0A182UPK0 A0A1S4GAQ9 A0A212FI49 E2BBN1 A0A3Q0JKY0 A0A195BVQ0 A0A182IBH9 A0A182X5U6 A0A182RK29 A0NFK2 A0A026W6I4 A0A084VSN0 A0A182MPH9 A0A3L8DSL2 A0A182TKB1 E0VQ13 A0A158N921 A0A2H1WU64 A0A2S2Q2I9 A0A154PIL3 A0A182W618 A0A2A4K4N0 A0A0L7QRK4 A0A2H8TMT1 A0A182RVN5 A0A0L0BQE5 A0A1B0CRC5 A0A195FUG6 A0A1B0FJ66 A0A1A9Z437 B4LQZ3 A0A1B0BQH0 A0A2H1W1K5 A0A182PTH7 A0A0L0C9B6 F4WHI3 A0A2M4CNM6 A0A2A4JL61 A0A182J2A1 A0A1I8MHH7 A0A0J7L7S8 A0A2M4BMW1 A0A1B0FFM0 A0A2A4JJQ6 A0A1I8NHV3 A0A3B0JU01 A0A0Q9X6E4 A0A3B0JWZ3 A0A1I8NN68
A0A2A4K0S7 A0A2W1BJM5 A0A2A4K0W3 A0A2A4JZ35 A0A194Q5K0 A0A212F656 A0A194RTL3 K7J3Y4 A0A1Y1LUS4 A0A0N0BBD3 A0A088AJG9 E2AYG8 A0A195F292 A0A3L8DWQ2 E2BDH1 F4X3S8 A0A026WPA9 A0A195DZY3 A0A067R5S2 A0A151XJQ1 A0A195BGK8 A0A195CRF6 A0A2A3E6U9 A0A088ALP5 A0A1J1IRA3 E2ACI2 A0A0L7QPA4 A0A182SH50 N6TI71 A0A0M9A176 A0A139WP45 A0A182YRZ5 A0A1B0GM78 A0A310SMD6 A0A194PUH5 A0A336LZ60 A0A182WFE3 A0A151J7V8 A0A154P8T4 A0A151I737 A0A182NGR2 K7IZ51 A0A151WP42 A0A182QJ96 A0A182UPK0 A0A1S4GAQ9 A0A212FI49 E2BBN1 A0A3Q0JKY0 A0A195BVQ0 A0A182IBH9 A0A182X5U6 A0A182RK29 A0NFK2 A0A026W6I4 A0A084VSN0 A0A182MPH9 A0A3L8DSL2 A0A182TKB1 E0VQ13 A0A158N921 A0A2H1WU64 A0A2S2Q2I9 A0A154PIL3 A0A182W618 A0A2A4K4N0 A0A0L7QRK4 A0A2H8TMT1 A0A182RVN5 A0A0L0BQE5 A0A1B0CRC5 A0A195FUG6 A0A1B0FJ66 A0A1A9Z437 B4LQZ3 A0A1B0BQH0 A0A2H1W1K5 A0A182PTH7 A0A0L0C9B6 F4WHI3 A0A2M4CNM6 A0A2A4JL61 A0A182J2A1 A0A1I8MHH7 A0A0J7L7S8 A0A2M4BMW1 A0A1B0FFM0 A0A2A4JJQ6 A0A1I8NHV3 A0A3B0JU01 A0A0Q9X6E4 A0A3B0JWZ3 A0A1I8NN68
PDB
6EFY
E-value=5.93109e-21,
Score=242
Ontologies
GO
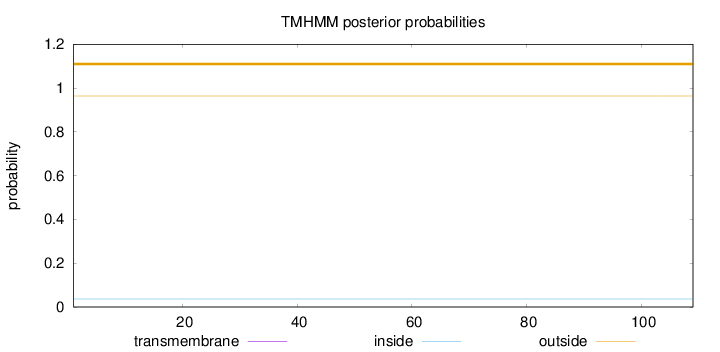
Topology
Length:
109
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00155
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03621
outside
1 - 109
Population Genetic Test Statistics
Pi
239.15994
Theta
180.935073
Tajima's D
0.777547
CLR
0.379386
CSRT
0.599970001499925
Interpretation
Uncertain
