Pre Gene Modal
BGIBMGA006357
Annotation
PREDICTED:_probable_ATP-dependent_RNA_helicase_pitchoune_[Papilio_polytes]
Full name
RNA helicase
+ More
Probable ATP-dependent RNA helicase pitchoune
Probable ATP-dependent RNA helicase pitchoune
Location in the cell
Nuclear Reliability : 3.608
Sequence
CDS
ATGCCGACACCAGATAAAATACTTCTCAGAAAAATAAGAAAACGAGAAAAACGAAAACTGAAACTTACATCTCAAAAAACTGATTCAAATCCTGAACCAGAAAAAGATATTGAAGCATTAAATAATGAAAAAGAAGCTAAGAAGAGAGTTGTACAAGACGGTGACACAAATGGAAAAGTTCCCAAAAAAAAATTGAAAAAGGTTAAAGAAGAGTTAAAAGAAGAACCAGTTGAAGTAAAATCTGAGGAAGAATCTAGTGATGACTCTCAAGATGAGCAAGACACTAAAGAAGATGATTCGGAAAAAAAATCGAACAACGATTTACCTGGATCAAGCCTCTGCTTAGGTATACTATCAGATCAAAAATTCACTGCACTGGAAGGCACAGTGTGCGAACCAACTCTTTTAGGAATTAAAGATATGGGCTTTATAACGATGACAGAGATTCAAGCTAAAGCTATACCTCCTTTGTTGGAAGGCAGGGATCTTGTGGGTGCTGCCAAAACTGGCTCTGGAAAAACACTAGCATTCTTAATACCGGCTATAGACCTTATATACAAATTAAAATTTAAACCAAGAAATGGTACTGGAGTCATCATTTTGTCACCCACAAGAGAGTTATCAATGCAAACATTTGGCGTTCTCATGGAATTGATGAAATACCACCACCATACATATGGTCTGGTAATGGGTGGTGCAAACAGAAGTACTGAAGCTCAGAAACTCTCTAAAGGTATCAACATTTTAGTTGCTACTCCTGGACGTTTACTGGATCACTTGCAAAATACACCAGACTTCTTGTATAAGAACCTTCAGTGCTTAGTTATTGATGAGGCTGACAGGATATTGGAAATTGGATTTGAAGAAGAAGTTAAACAGATCATAAAATTGCTGCCAAAACGCCGGCAAACTATGTTATTCAGTGCTACACAAACAAAGAAGACTGAAGCACTAACTGCTTTGGCATTGAAGCATGAACCCGTGTATGTTGGGGTTGATGACCACAGAGAACAGGCAACTGTTGATAGTTTAGAACAGGGATATATCGTATGCCCATCAGAGAAACGTATGATGGTTTTATTCACATTCCTTAAGAAGAATAGAAAAAAGAAGGTTATGGTATTCTTTTCAACCTGTATGTCTGTCAAATACCACCATGAACTTTTCAATTATATTGACCTTCCTGTGATGTCCATACATGGAAAACAACAACAAACGAAGCGTACAACAACATTCTTCCAGTTTTGCAATGCTGAATCTGGTATACTACTTTGTACTGATGTCGCGGCGCGTGGTCTGGACATTCCGGCCGTTGATTGGATTGTACAGTATGATCCTCCAGATGATCCAAAGGAATACATCCATCGGGTTGGAAGAACTGCGAGGGGTCTAGGAACTAGTGGACACGCTTTGTTATTCTTACGTCCAGAAGAGCTTGGATTCTTACGATACTTAAAGCAGTCGAGAGTTACCCTAAACGAATTTGAATTCTCATGGAATAAAGTTGCAGATATACAACTACAGCTAGAAAAACTTATCTCTCGAAACTATTTCCTGAATCAATCTGCAAAAGAAGCCTTCAAAAGTTACCTGAGAGCCTATGATTCTCATCACTTGAAAACGATTTTTGACATTGATACAATCGATTTAGCTAAGGCATCGAAATCATTTGGTTTCAATGTACCGCCCGCTGTAGAACTAAAAGTCGCAAACAAAGGACCTCCACAGAAAAGAAAGGGAGGTGGTGGTTATGGATACTTCAAGTCCTTAAACGCTCCATCCGATTTCAAAAGGAAGACAGATAAAACGAAAATATACAGACAAAAGGGGAACAATAAGAAAACGATCAGTTGA
Protein
MPTPDKILLRKIRKREKRKLKLTSQKTDSNPEPEKDIEALNNEKEAKKRVVQDGDTNGKVPKKKLKKVKEELKEEPVEVKSEEESSDDSQDEQDTKEDDSEKKSNNDLPGSSLCLGILSDQKFTALEGTVCEPTLLGIKDMGFITMTEIQAKAIPPLLEGRDLVGAAKTGSGKTLAFLIPAIDLIYKLKFKPRNGTGVIILSPTRELSMQTFGVLMELMKYHHHTYGLVMGGANRSTEAQKLSKGINILVATPGRLLDHLQNTPDFLYKNLQCLVIDEADRILEIGFEEEVKQIIKLLPKRRQTMLFSATQTKKTEALTALALKHEPVYVGVDDHREQATVDSLEQGYIVCPSEKRMMVLFTFLKKNRKKKVMVFFSTCMSVKYHHELFNYIDLPVMSIHGKQQQTKRTTTFFQFCNAESGILLCTDVAARGLDIPAVDWIVQYDPPDDPKEYIHRVGRTARGLGTSGHALLFLRPEELGFLRYLKQSRVTLNEFEFSWNKVADIQLQLEKLISRNYFLNQSAKEAFKSYLRAYDSHHLKTIFDIDTIDLAKASKSFGFNVPPAVELKVANKGPPQKRKGGGGYGYFKSLNAPSDFKRKTDKTKIYRQKGNNKKTIS
Summary
Description
Probable RNA-dependent helicase. Functions in cell growth and proliferation. May have a role in ribosome biogenesis and, consequently, in protein biosynthesis.
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Miscellaneous
Mutation in the pit gene produce larvae that cannot grow beyond the first instar larval stage although they can live as long as 7-10 days. All the tissues are equally affected and the perfectly shaped larvae are indistinguishable from first instar wild-type animals.
'Pitchoune' means 'small' in Provence (Southern part of France).
'Pitchoune' means 'small' in Provence (Southern part of France).
Similarity
Belongs to the DEAD box helicase family.
Belongs to the DEAD box helicase family. DDX18/HAS1 subfamily.
Belongs to the DEAD box helicase family. DDX18/HAS1 subfamily.
Keywords
ATP-binding
Complete proteome
Helicase
Hydrolase
Nucleotide-binding
Nucleus
Reference proteome
RNA-binding
Feature
chain Probable ATP-dependent RNA helicase pitchoune
Uniprot
A0A2W1BNP3
A0A2A4J767
A0A212F650
S4PLI4
H9JA12
A0A194PXI2
+ More
A0A0L7LBX8 D6W8J8 A0A1W4XJB7 A0A0T6BH44 K7J159 A0A1Y1LJF9 A0A2J7RRR1 A0A1B6E4Q7 A0A0N0U4N8 A0A195FFG3 A0A158NH79 K7J154 A0A0C9R1I3 E9IJQ2 A0A0K8TNZ2 A0A195B6U5 A0A195DXU2 A0A154P607 E0VZA3 A0A1I8N0G9 A0A026X1E9 A0A146MF85 N6UM89 U4U8K9 W8BRJ4 A0A2S2ND42 A0A2H8TDD5 A0A1I8Q5V6 A0A336LY65 A0A2S2QMA8 A0A1B0G9E6 A0A0L0CEK4 A0A1B0A332 A0A3Q0IP60 B4HM85 A0A0B4LHF7 B3LVL5 B3P7N7 Q9VD51 B4K6L4 X1WV14 B4PLK0 A0A0K8TZH5 B5DW65 A0A034V7T4 A0A0Q9X2B0 A0A0Q9X1W3 B4M0M9 A0A3B0K2U9 A0A067QLN7 A0A0Q9WRB4 A0A0M4EGD0 A0A3L8DQJ5 A0A1W4W1L2 A0A1A9VGC8 B4JRL9 A0A0L7QJS7 A0A151XD17 B4NHR6 A0A1B6GIA3 T1PH19 A0A1J1J3T6 A0A0N8EL32 A0A0P5I9A7 A0A0P6GGE3 A0A0N8EAF2 A0A0P6HAY4 A0A0P5BTJ0 A0A0P5IMI1 A0A0P5N0P0 A0A0P5SDG0 A0A0P5PMA1 A0A0P5NRZ2 A0A0P5B2M3 A0A0P4ZEE9 A0A0P5D3X7 A0A0P5PEL9 A0A0P5A3H4 A0A0P5MHR0 A0A0P5NCH5 A0A0P5S6M3 A0A0N8B4B4 A0A0P6DL16 A0A0P5PQS6 A0A0P5UPV5 A0A0P5Q0N6 A0A0P5R2F2 A0A0P5HRP9 A0A0P5RD11 A0A0P5VBT1 A0A0P6D864 A0A023F4R2 A0A0P5YTJ8 A0A0P5LB25
A0A0L7LBX8 D6W8J8 A0A1W4XJB7 A0A0T6BH44 K7J159 A0A1Y1LJF9 A0A2J7RRR1 A0A1B6E4Q7 A0A0N0U4N8 A0A195FFG3 A0A158NH79 K7J154 A0A0C9R1I3 E9IJQ2 A0A0K8TNZ2 A0A195B6U5 A0A195DXU2 A0A154P607 E0VZA3 A0A1I8N0G9 A0A026X1E9 A0A146MF85 N6UM89 U4U8K9 W8BRJ4 A0A2S2ND42 A0A2H8TDD5 A0A1I8Q5V6 A0A336LY65 A0A2S2QMA8 A0A1B0G9E6 A0A0L0CEK4 A0A1B0A332 A0A3Q0IP60 B4HM85 A0A0B4LHF7 B3LVL5 B3P7N7 Q9VD51 B4K6L4 X1WV14 B4PLK0 A0A0K8TZH5 B5DW65 A0A034V7T4 A0A0Q9X2B0 A0A0Q9X1W3 B4M0M9 A0A3B0K2U9 A0A067QLN7 A0A0Q9WRB4 A0A0M4EGD0 A0A3L8DQJ5 A0A1W4W1L2 A0A1A9VGC8 B4JRL9 A0A0L7QJS7 A0A151XD17 B4NHR6 A0A1B6GIA3 T1PH19 A0A1J1J3T6 A0A0N8EL32 A0A0P5I9A7 A0A0P6GGE3 A0A0N8EAF2 A0A0P6HAY4 A0A0P5BTJ0 A0A0P5IMI1 A0A0P5N0P0 A0A0P5SDG0 A0A0P5PMA1 A0A0P5NRZ2 A0A0P5B2M3 A0A0P4ZEE9 A0A0P5D3X7 A0A0P5PEL9 A0A0P5A3H4 A0A0P5MHR0 A0A0P5NCH5 A0A0P5S6M3 A0A0N8B4B4 A0A0P6DL16 A0A0P5PQS6 A0A0P5UPV5 A0A0P5Q0N6 A0A0P5R2F2 A0A0P5HRP9 A0A0P5RD11 A0A0P5VBT1 A0A0P6D864 A0A023F4R2 A0A0P5YTJ8 A0A0P5LB25
EC Number
3.6.4.13
Pubmed
28756777
22118469
23622113
19121390
26354079
26227816
+ More
18362917 19820115 20075255 28004739 21347285 21282665 26369729 20566863 25315136 24508170 26823975 23537049 24495485 26108605 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 9716523 12537569 17550304 15632085 25348373 24845553 30249741 25474469
18362917 19820115 20075255 28004739 21347285 21282665 26369729 20566863 25315136 24508170 26823975 23537049 24495485 26108605 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 9716523 12537569 17550304 15632085 25348373 24845553 30249741 25474469
EMBL
KZ150004
PZC75254.1
NWSH01002631
PCG67801.1
AGBW02010074
OWR49215.1
+ More
GAIX01004010 JAA88550.1 BABH01009406 KQ459589 KPI97469.1 JTDY01001746 KOB73007.1 KQ971312 EEZ98368.1 LJIG01000276 KRT86664.1 GEZM01055754 JAV72948.1 NEVH01000599 PNF43502.1 GEDC01004386 JAS32912.1 KQ435824 KOX72228.1 KQ981625 KYN39123.1 ADTU01015209 GBYB01001815 JAG71582.1 GL763877 EFZ19207.1 GDAI01001998 JAI15605.1 KQ976574 KYM80238.1 KQ980107 KYN17693.1 KQ434813 KZC06784.1 DS235851 EEB18709.1 KK107063 EZA61209.1 GDHC01000957 JAQ17672.1 APGK01017820 KB740053 ENN81801.1 KB632093 ERL88693.1 GAMC01007082 JAB99473.1 GGMR01002494 MBY15113.1 GFXV01000294 MBW12099.1 UFQT01000292 SSX22905.1 GGMS01009625 MBY78828.1 CCAG010017460 JRES01000501 KNC30690.1 CH480815 EDW43133.1 AE014297 AHN57458.1 CH902617 EDV42585.1 CH954182 EDV54198.1 U84552 AY071402 AY119620 CH933806 EDW16314.1 ABLF02029216 CM000160 EDW97949.1 GDHF01032633 JAI19681.1 CM000070 EDY68110.1 GAKP01021350 GAKP01021349 JAC37603.1 KRG02045.1 KRG02044.1 CH940650 EDW67321.1 OUUW01000005 SPP80279.1 KK853180 KDR10194.1 KRF83246.1 CP012526 ALC47404.1 QOIP01000005 RLU22532.1 CH916373 EDV94409.1 KQ415041 KOC58877.1 KQ982294 KYQ58210.1 CH964272 EDW84676.1 GECZ01007597 JAS62172.1 KA648066 AFP62695.1 CVRI01000070 CRL07099.1 GDIQ01016212 JAN78525.1 GDIQ01215846 GDIQ01215845 GDIQ01110641 JAK35880.1 GDIQ01044008 JAN50729.1 GDIQ01046052 JAN48685.1 GDIQ01044007 JAN50730.1 GDIP01180278 JAJ43124.1 GDIQ01211896 JAK39829.1 GDIQ01160109 JAK91616.1 GDIQ01095090 JAL56636.1 GDIQ01136922 JAL14804.1 GDIQ01160108 JAK91617.1 GDIP01195228 JAJ28174.1 GDIP01218079 JAJ05323.1 GDIP01161528 GDIQ01064054 JAJ61874.1 JAN30683.1 GDIQ01129677 LRGB01000687 JAL22049.1 KZS17013.1 GDIP01208028 JAJ15374.1 GDIQ01158013 JAK93712.1 GDIQ01147178 JAL04548.1 GDIQ01108734 JAL42992.1 GDIQ01214147 JAK37578.1 GDIQ01075947 JAN18790.1 GDIQ01145826 JAL05900.1 GDIP01110037 JAL93677.1 GDIQ01120777 JAL30949.1 GDIQ01108733 JAL42993.1 GDIQ01229241 JAK22484.1 GDIQ01108732 JAL42994.1 GDIP01102489 JAM01226.1 GDIQ01081893 JAN12844.1 GBBI01002310 JAC16402.1 GDIP01053657 JAM50058.1 GDIQ01176847 JAK74878.1
GAIX01004010 JAA88550.1 BABH01009406 KQ459589 KPI97469.1 JTDY01001746 KOB73007.1 KQ971312 EEZ98368.1 LJIG01000276 KRT86664.1 GEZM01055754 JAV72948.1 NEVH01000599 PNF43502.1 GEDC01004386 JAS32912.1 KQ435824 KOX72228.1 KQ981625 KYN39123.1 ADTU01015209 GBYB01001815 JAG71582.1 GL763877 EFZ19207.1 GDAI01001998 JAI15605.1 KQ976574 KYM80238.1 KQ980107 KYN17693.1 KQ434813 KZC06784.1 DS235851 EEB18709.1 KK107063 EZA61209.1 GDHC01000957 JAQ17672.1 APGK01017820 KB740053 ENN81801.1 KB632093 ERL88693.1 GAMC01007082 JAB99473.1 GGMR01002494 MBY15113.1 GFXV01000294 MBW12099.1 UFQT01000292 SSX22905.1 GGMS01009625 MBY78828.1 CCAG010017460 JRES01000501 KNC30690.1 CH480815 EDW43133.1 AE014297 AHN57458.1 CH902617 EDV42585.1 CH954182 EDV54198.1 U84552 AY071402 AY119620 CH933806 EDW16314.1 ABLF02029216 CM000160 EDW97949.1 GDHF01032633 JAI19681.1 CM000070 EDY68110.1 GAKP01021350 GAKP01021349 JAC37603.1 KRG02045.1 KRG02044.1 CH940650 EDW67321.1 OUUW01000005 SPP80279.1 KK853180 KDR10194.1 KRF83246.1 CP012526 ALC47404.1 QOIP01000005 RLU22532.1 CH916373 EDV94409.1 KQ415041 KOC58877.1 KQ982294 KYQ58210.1 CH964272 EDW84676.1 GECZ01007597 JAS62172.1 KA648066 AFP62695.1 CVRI01000070 CRL07099.1 GDIQ01016212 JAN78525.1 GDIQ01215846 GDIQ01215845 GDIQ01110641 JAK35880.1 GDIQ01044008 JAN50729.1 GDIQ01046052 JAN48685.1 GDIQ01044007 JAN50730.1 GDIP01180278 JAJ43124.1 GDIQ01211896 JAK39829.1 GDIQ01160109 JAK91616.1 GDIQ01095090 JAL56636.1 GDIQ01136922 JAL14804.1 GDIQ01160108 JAK91617.1 GDIP01195228 JAJ28174.1 GDIP01218079 JAJ05323.1 GDIP01161528 GDIQ01064054 JAJ61874.1 JAN30683.1 GDIQ01129677 LRGB01000687 JAL22049.1 KZS17013.1 GDIP01208028 JAJ15374.1 GDIQ01158013 JAK93712.1 GDIQ01147178 JAL04548.1 GDIQ01108734 JAL42992.1 GDIQ01214147 JAK37578.1 GDIQ01075947 JAN18790.1 GDIQ01145826 JAL05900.1 GDIP01110037 JAL93677.1 GDIQ01120777 JAL30949.1 GDIQ01108733 JAL42993.1 GDIQ01229241 JAK22484.1 GDIQ01108732 JAL42994.1 GDIP01102489 JAM01226.1 GDIQ01081893 JAN12844.1 GBBI01002310 JAC16402.1 GDIP01053657 JAM50058.1 GDIQ01176847 JAK74878.1
Proteomes
UP000218220
UP000007151
UP000005204
UP000053268
UP000037510
UP000007266
+ More
UP000192223 UP000002358 UP000235965 UP000053105 UP000078541 UP000005205 UP000078540 UP000078492 UP000076502 UP000009046 UP000095301 UP000053097 UP000019118 UP000030742 UP000095300 UP000092444 UP000037069 UP000092445 UP000079169 UP000001292 UP000000803 UP000007801 UP000008711 UP000009192 UP000007819 UP000002282 UP000001819 UP000008792 UP000268350 UP000027135 UP000092553 UP000279307 UP000192221 UP000078200 UP000001070 UP000053825 UP000075809 UP000007798 UP000183832 UP000076858
UP000192223 UP000002358 UP000235965 UP000053105 UP000078541 UP000005205 UP000078540 UP000078492 UP000076502 UP000009046 UP000095301 UP000053097 UP000019118 UP000030742 UP000095300 UP000092444 UP000037069 UP000092445 UP000079169 UP000001292 UP000000803 UP000007801 UP000008711 UP000009192 UP000007819 UP000002282 UP000001819 UP000008792 UP000268350 UP000027135 UP000092553 UP000279307 UP000192221 UP000078200 UP000001070 UP000053825 UP000075809 UP000007798 UP000183832 UP000076858
Pfam
Interpro
IPR000629
RNA-helicase_DEAD-box_CS
+ More
IPR014014 RNA_helicase_DEAD_Q_motif
IPR011545 DEAD/DEAH_box_helicase_dom
IPR025313 DUF4217
IPR027417 P-loop_NTPase
IPR001650 Helicase_C
IPR014001 Helicase_ATP-bd
IPR019049 Nucleoporin_prot_Ndc1/Nup
IPR036612 KH_dom_type_1_sf
IPR004088 KH_dom_type_1
IPR032377 STAR_dimer
IPR004087 KH_dom
IPR014014 RNA_helicase_DEAD_Q_motif
IPR011545 DEAD/DEAH_box_helicase_dom
IPR025313 DUF4217
IPR027417 P-loop_NTPase
IPR001650 Helicase_C
IPR014001 Helicase_ATP-bd
IPR019049 Nucleoporin_prot_Ndc1/Nup
IPR036612 KH_dom_type_1_sf
IPR004088 KH_dom_type_1
IPR032377 STAR_dimer
IPR004087 KH_dom
Gene 3D
CDD
ProteinModelPortal
A0A2W1BNP3
A0A2A4J767
A0A212F650
S4PLI4
H9JA12
A0A194PXI2
+ More
A0A0L7LBX8 D6W8J8 A0A1W4XJB7 A0A0T6BH44 K7J159 A0A1Y1LJF9 A0A2J7RRR1 A0A1B6E4Q7 A0A0N0U4N8 A0A195FFG3 A0A158NH79 K7J154 A0A0C9R1I3 E9IJQ2 A0A0K8TNZ2 A0A195B6U5 A0A195DXU2 A0A154P607 E0VZA3 A0A1I8N0G9 A0A026X1E9 A0A146MF85 N6UM89 U4U8K9 W8BRJ4 A0A2S2ND42 A0A2H8TDD5 A0A1I8Q5V6 A0A336LY65 A0A2S2QMA8 A0A1B0G9E6 A0A0L0CEK4 A0A1B0A332 A0A3Q0IP60 B4HM85 A0A0B4LHF7 B3LVL5 B3P7N7 Q9VD51 B4K6L4 X1WV14 B4PLK0 A0A0K8TZH5 B5DW65 A0A034V7T4 A0A0Q9X2B0 A0A0Q9X1W3 B4M0M9 A0A3B0K2U9 A0A067QLN7 A0A0Q9WRB4 A0A0M4EGD0 A0A3L8DQJ5 A0A1W4W1L2 A0A1A9VGC8 B4JRL9 A0A0L7QJS7 A0A151XD17 B4NHR6 A0A1B6GIA3 T1PH19 A0A1J1J3T6 A0A0N8EL32 A0A0P5I9A7 A0A0P6GGE3 A0A0N8EAF2 A0A0P6HAY4 A0A0P5BTJ0 A0A0P5IMI1 A0A0P5N0P0 A0A0P5SDG0 A0A0P5PMA1 A0A0P5NRZ2 A0A0P5B2M3 A0A0P4ZEE9 A0A0P5D3X7 A0A0P5PEL9 A0A0P5A3H4 A0A0P5MHR0 A0A0P5NCH5 A0A0P5S6M3 A0A0N8B4B4 A0A0P6DL16 A0A0P5PQS6 A0A0P5UPV5 A0A0P5Q0N6 A0A0P5R2F2 A0A0P5HRP9 A0A0P5RD11 A0A0P5VBT1 A0A0P6D864 A0A023F4R2 A0A0P5YTJ8 A0A0P5LB25
A0A0L7LBX8 D6W8J8 A0A1W4XJB7 A0A0T6BH44 K7J159 A0A1Y1LJF9 A0A2J7RRR1 A0A1B6E4Q7 A0A0N0U4N8 A0A195FFG3 A0A158NH79 K7J154 A0A0C9R1I3 E9IJQ2 A0A0K8TNZ2 A0A195B6U5 A0A195DXU2 A0A154P607 E0VZA3 A0A1I8N0G9 A0A026X1E9 A0A146MF85 N6UM89 U4U8K9 W8BRJ4 A0A2S2ND42 A0A2H8TDD5 A0A1I8Q5V6 A0A336LY65 A0A2S2QMA8 A0A1B0G9E6 A0A0L0CEK4 A0A1B0A332 A0A3Q0IP60 B4HM85 A0A0B4LHF7 B3LVL5 B3P7N7 Q9VD51 B4K6L4 X1WV14 B4PLK0 A0A0K8TZH5 B5DW65 A0A034V7T4 A0A0Q9X2B0 A0A0Q9X1W3 B4M0M9 A0A3B0K2U9 A0A067QLN7 A0A0Q9WRB4 A0A0M4EGD0 A0A3L8DQJ5 A0A1W4W1L2 A0A1A9VGC8 B4JRL9 A0A0L7QJS7 A0A151XD17 B4NHR6 A0A1B6GIA3 T1PH19 A0A1J1J3T6 A0A0N8EL32 A0A0P5I9A7 A0A0P6GGE3 A0A0N8EAF2 A0A0P6HAY4 A0A0P5BTJ0 A0A0P5IMI1 A0A0P5N0P0 A0A0P5SDG0 A0A0P5PMA1 A0A0P5NRZ2 A0A0P5B2M3 A0A0P4ZEE9 A0A0P5D3X7 A0A0P5PEL9 A0A0P5A3H4 A0A0P5MHR0 A0A0P5NCH5 A0A0P5S6M3 A0A0N8B4B4 A0A0P6DL16 A0A0P5PQS6 A0A0P5UPV5 A0A0P5Q0N6 A0A0P5R2F2 A0A0P5HRP9 A0A0P5RD11 A0A0P5VBT1 A0A0P6D864 A0A023F4R2 A0A0P5YTJ8 A0A0P5LB25
PDB
6EM5
E-value=1.09598e-169,
Score=1533
Ontologies
GO
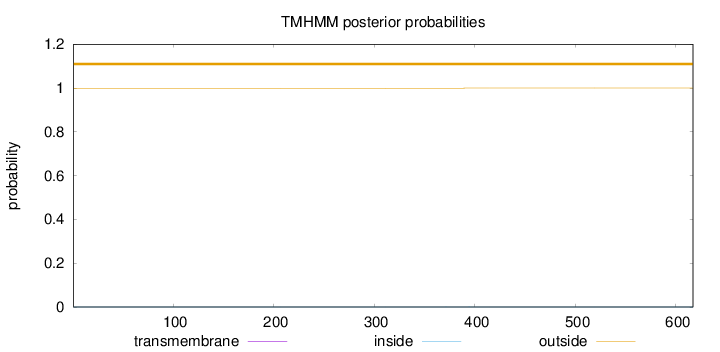
Topology
Subcellular location
Length:
617
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00504999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00050
outside
1 - 617
Population Genetic Test Statistics
Pi
234.21002
Theta
156.151962
Tajima's D
1.466759
CLR
0
CSRT
0.778361081945903
Interpretation
Uncertain
