Gene
KWMTBOMO03473 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006523
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_glucosidase_2_subunit_beta-like_[Bombyx_mori]
Full name
Glucosidase 2 subunit beta
Alternative Name
80K-H protein
Glucosidase II subunit beta
Protein kinase C substrate 60.1 kDa protein heavy chain
Glucosidase II subunit beta
Protein kinase C substrate 60.1 kDa protein heavy chain
Location in the cell
Cytoplasmic Reliability : 1.617 Nuclear Reliability : 2.209
Sequence
CDS
ATGGTCTGTAAAACATGGATCGACCGATTTTCCTCCTATTTCGTAATATTTTGTTCCGTCATCATATTTGCCCAGTCCGATGTGCCTAGACCCCGCGGTGTATCTTTGTCTAAAGCATCTCTCTATTTACCGACAAAAGATTTTACTTGTTTTGATGGAACTGCTACAATACCTTTCAGCTACGTTAATGACGACTACTGTGATTGTTTCGACGGTAGCGATGAACCCGGTACATCGGCTTGTATAAACGGTGTTTTCCATTGTACAAATGCCGGACACCGGCCTCAGAATTTGCCGAGTTCCAGGGTTAATGACGGAGTCTGTGATTGCTGTGATGGAACAGATGAATACGCTAATCCTACGGCCTGTACTAACATTTGTGAAGAGCTCGGAAAAGAAGCCCGAGCAGAAGCTCAAAGAGTAGCAGAGTTGCATAAAGCAGGCAGCCAATTGAGAATAGACCTCATAGAAAAAGGAAATAAGAAGCGAAATGAAATGGCAGAACAACTAACACAACTTGAAAAAGACAAATCTGAAGCAGAAAAAATAAAGGCTGAAAAAGAGCTTCTAAAGAATGATCTAGAAATGAAAGAAAATGAGGTTTTGAAAGTATATAGAGAAGCAGAAGAATTGGAAAAACAAAAGAAATTAGACCAAGAGAAAGAAACTAATTTAAAAGAATCAACGGAACACTTTAATAGATTTGATTCAAATAATGATGGAGAGTTAAGTATAGATGAAATCAAAGTTGTTAATGTATTTGATAGGAACAAGGATGGCCAAGTTGATGATGATGAAGTAAAGATGTTCTTGGGTGGTGATGAAAAAGTAGATAAAGATGCATTCTTGTCCACAACTTGGCCTATCTTAAAACCACTTTTGATGATGGAGCAAGGTATGTTTAAACCAGAGGATGCTAAAACAGATACCACGGAGACAGAAGCTGAAGAAAACCATGAAGAATTGGATGCTGATTTTGAAGGAAATGAAGAAGATAATAGCGATTTAGATTTACAAGAAGAACATGAAGAAGATGAGACTGTGTCAGAGCAGTCAAAATCATATAATGATGAGACTCAAAAAATTGTTGATCAGGCTTCAGAAGCTCGCAGGCAATACACAGATGCTGAGCGAACTGTTAGAGAAATAGAGTCTAACATTAGAAACATAAAACAGAACTTAGAAAAGGATTATGGTCTAGAACAGGAATTTGCTTCCTTGGATGGTGATTGCTTTGAATATGAAGACAAAGAATATGTATACAAACTGTGTATGTTCCAGAAGGTAACACAAAAATCCAAGAATGGTGGTATGGAGGTAGGTCTTGGTAATTGGGGAGAATGGGCAGGTCCTGAAAACAATAAATACTCTGTGATGAAATACACAAATGGAATTGCTTGCTGGAATGGTCCAAGTCGAACAACAACTGTTAATGTCAACTGTGACTTGGAAACAAAGATCACATCAGTAACTGAACCTTTCCGTTGTGAATACAAAATGGAATTAAGCACTCCTGCTGCATGTGATGATTCTAGCACATCACAAGAGCATACATCACATGATGAACTTTAA
Protein
MVCKTWIDRFSSYFVIFCSVIIFAQSDVPRPRGVSLSKASLYLPTKDFTCFDGTATIPFSYVNDDYCDCFDGSDEPGTSACINGVFHCTNAGHRPQNLPSSRVNDGVCDCCDGTDEYANPTACTNICEELGKEARAEAQRVAELHKAGSQLRIDLIEKGNKKRNEMAEQLTQLEKDKSEAEKIKAEKELLKNDLEMKENEVLKVYREAEELEKQKKLDQEKETNLKESTEHFNRFDSNNDGELSIDEIKVVNVFDRNKDGQVDDDEVKMFLGGDEKVDKDAFLSTTWPILKPLLMMEQGMFKPEDAKTDTTETEAEENHEELDADFEGNEEDNSDLDLQEEHEEDETVSEQSKSYNDETQKIVDQASEARRQYTDAERTVREIESNIRNIKQNLEKDYGLEQEFASLDGDCFEYEDKEYVYKLCMFQKVTQKSKNGGMEVGLGNWGEWAGPENNKYSVMKYTNGIACWNGPSRTTTVNVNCDLETKITSVTEPFRCEYKMELSTPAACDDSSTSQEHTSHDEL
Summary
Description
Regulatory subunit of glucosidase II that cleaves sequentially the 2 innermost alpha-1,3-linked glucose residues from the Glc(2)Man(9)GlcNAc(2) oligosaccharide precursor of immature glycoproteins (PubMed:27462106, PubMed:9148925). Required for efficient PKD1/Polycystin-1 biogenesis and trafficking to the plasma membrane of the primary cilia (PubMed:21685914).
Subunit
Heterodimer of a catalytic alpha subunit (GANAB) and a beta subunit (PRKCSH) (PubMed:9148925, PubMed:27462106). Binds glycosylated PTPRC (PubMed:9148925).
Keywords
3D-structure
Alternative splicing
Calcium
Complete proteome
Direct protein sequencing
Disulfide bond
Endoplasmic reticulum
Glycoprotein
Metal-binding
Phosphoprotein
Reference proteome
Repeat
Signal
Feature
chain Glucosidase 2 subunit beta
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
L8B6C5
A0A1E1WUQ0
A0A345W862
A0A194PVQ6
A0A194RQ52
S4PCP7
+ More
A0A2A4J8T2 A0A2W1BRF2 A0A212F655 A0A2H1VSG9 A0A0L7LAG7 I4DNV0 D6W9R4 A0A1Y1M7I2 V5G2I9 A0A1B6D3J4 A0A1B0CR59 A0A1L8DJ45 A0A1L8DJ39 A0A1L8DM68 A0A1L8DM51 A0A0N1IT49 A0A1Q3FF99 A0A2P8XVP6 A0A0P4ZG66 A0A0P4YW50 A0A1D2N278 A0A088AUN3 A0A0P4WUM2 A0A0L7R7R0 A0A0P5Z9C2 A0A0P5RRM1 A0A0P5Y5P3 A0A0P5IT85 A0A0J7KXH9 E9G8J3 A0A2A3ER50 A0A182IWR0 U4TZH6 N6T690 E9IKT6 A0A154PSW3 A0A195FCW2 E2AQZ0 A0A195EL25 A0A158NZI4 A0A0P6I4Q2 A0A023EUW9 B0WS45 A0A151WR87 J9JPV0 A0A2H8TH21 A0A0P5DIB6 A0A0P6BRN7 A0A195CJ66 K7J4G8 A0A3L8D713 F4WL80 A0A026W1M5 B3MMH5 B4N1C4 B4I5B3 A0A1A9WM41 A0A0J9TQ58 Q9VJD1 A0A1B6GD55 B4P8R1 A0A132A4G4 Q29LB3 B4GQB0 A0A1A9Y5B2 A0A1S3JT06 B3NMG7 A0A1J1HJU0 A0A1B0A3X9 A0A0A1XJ19 A0A3B0JCE6 W8BAL3 R4WR17 A0A310TPI7 A0A1W4ULN9 Q6AZH1 A0A2S2Q9G1 A0A034WM66 A0A0N8JZG3 Q6GLZ2 A0A1B0FH60 A0A131XVY0 A0A3N0YY10 A8WGX5 Q6P373 O08795-2 A0A0K8VHR2 A0A0P6J571 G3UJW5 H0XEE9 A0A2K5WQ22 A0A0P5CQM8 F7C6C6
A0A2A4J8T2 A0A2W1BRF2 A0A212F655 A0A2H1VSG9 A0A0L7LAG7 I4DNV0 D6W9R4 A0A1Y1M7I2 V5G2I9 A0A1B6D3J4 A0A1B0CR59 A0A1L8DJ45 A0A1L8DJ39 A0A1L8DM68 A0A1L8DM51 A0A0N1IT49 A0A1Q3FF99 A0A2P8XVP6 A0A0P4ZG66 A0A0P4YW50 A0A1D2N278 A0A088AUN3 A0A0P4WUM2 A0A0L7R7R0 A0A0P5Z9C2 A0A0P5RRM1 A0A0P5Y5P3 A0A0P5IT85 A0A0J7KXH9 E9G8J3 A0A2A3ER50 A0A182IWR0 U4TZH6 N6T690 E9IKT6 A0A154PSW3 A0A195FCW2 E2AQZ0 A0A195EL25 A0A158NZI4 A0A0P6I4Q2 A0A023EUW9 B0WS45 A0A151WR87 J9JPV0 A0A2H8TH21 A0A0P5DIB6 A0A0P6BRN7 A0A195CJ66 K7J4G8 A0A3L8D713 F4WL80 A0A026W1M5 B3MMH5 B4N1C4 B4I5B3 A0A1A9WM41 A0A0J9TQ58 Q9VJD1 A0A1B6GD55 B4P8R1 A0A132A4G4 Q29LB3 B4GQB0 A0A1A9Y5B2 A0A1S3JT06 B3NMG7 A0A1J1HJU0 A0A1B0A3X9 A0A0A1XJ19 A0A3B0JCE6 W8BAL3 R4WR17 A0A310TPI7 A0A1W4ULN9 Q6AZH1 A0A2S2Q9G1 A0A034WM66 A0A0N8JZG3 Q6GLZ2 A0A1B0FH60 A0A131XVY0 A0A3N0YY10 A8WGX5 Q6P373 O08795-2 A0A0K8VHR2 A0A0P6J571 G3UJW5 H0XEE9 A0A2K5WQ22 A0A0P5CQM8 F7C6C6
Pubmed
23376632
29258441
26354079
23622113
28756777
22118469
+ More
26227816 22651552 18362917 19820115 28004739 29403074 27289101 21292972 23537049 21282665 20798317 21347285 24945155 20075255 30249741 21719571 24508170 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 26555130 15632085 25830018 24495485 23691247 25348373 9148925 15489334 21183079 21685914 23806337 27462106 17431167
26227816 22651552 18362917 19820115 28004739 29403074 27289101 21292972 23537049 21282665 20798317 21347285 24945155 20075255 30249741 21719571 24508170 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 26555130 15632085 25830018 24495485 23691247 25348373 9148925 15489334 21183079 21685914 23806337 27462106 17431167
EMBL
AB747259
BAM78679.1
GDQN01000387
JAT90667.1
MH071439
AXJ21600.1
+ More
KQ459589 KPI97471.1 KQ459896 KPJ19450.1 GAIX01004066 JAA88494.1 NWSH01002631 PCG67803.1 KZ150004 PZC75256.1 AGBW02010074 OWR49217.1 ODYU01004177 SOQ43767.1 JTDY01002064 KOB72196.1 AK403151 BAM19590.1 KQ971312 EEZ98119.2 GEZM01038259 JAV81683.1 GALX01004236 JAB64230.1 GEDC01017026 JAS20272.1 AJWK01024420 GFDF01007596 JAV06488.1 GFDF01007595 JAV06489.1 GFDF01006630 JAV07454.1 GFDF01006545 JAV07539.1 KQ435883 KOX69658.1 GFDL01008834 JAV26211.1 PYGN01001278 PSN36075.1 GDIP01221092 GDIP01213238 GDIP01137490 GDIP01130596 GDIP01130595 GDIP01127834 GDIP01113000 GDIP01061107 LRGB01002451 JAJ10164.1 KZS07500.1 GDIP01222238 GDIP01222237 GDIP01184635 GDIP01156603 GDIP01146489 GDIP01146488 GDIQ01136981 GDIP01111739 JAJ01165.1 JAL14745.1 LJIJ01000284 ODM99396.1 GDIP01251373 GDIP01221093 GDIP01140845 GDIP01140844 GDIP01137492 GDIP01137491 GDIP01130594 GDIP01113002 GDIP01113001 GDIP01074409 JAI72028.1 KQ414638 KOC66858.1 GDIP01222239 GDIP01184634 GDIP01115742 GDIP01111738 GDIP01085060 GDIP01073158 GDIP01065109 GDIP01047016 GDIP01047015 JAM56699.1 GDIQ01102850 JAL48876.1 GDIP01062603 JAM41112.1 GDIQ01208311 JAK43414.1 LBMM01002287 KMQ95026.1 GL732535 EFX84260.1 KZ288192 PBC34245.1 KB631903 ERL87019.1 APGK01042136 KB741002 ENN75729.1 GL764026 EFZ18732.1 KQ435075 KZC14414.1 KQ981685 KYN38037.1 GL441846 EFN64136.1 KQ978747 KYN28642.1 ADTU01004641 GDIQ01009391 JAN85346.1 GAPW01000816 JAC12782.1 DS232064 EDS33664.1 KQ982807 KYQ50376.1 ABLF02023169 GFXV01001525 MBW13330.1 GDIP01161692 JAJ61710.1 GDIP01025764 JAM77951.1 KQ977720 KYN00477.1 QOIP01000012 RLU16255.1 GL888207 EGI65043.1 KK107488 EZA49928.1 CH902620 EDV30921.1 CH963920 EDW78114.1 CH480822 EDW55569.1 CM002910 KMY90720.1 AE014134 AY058725 AAF53621.1 AAL13954.1 AFH03737.1 GECZ01009391 JAS60378.1 CM000158 EDW90169.1 JXLN01010490 KPM05814.1 CH379060 EAL34132.2 CH479187 EDW39782.1 CH954179 EDV54838.1 CVRI01000006 CRK88328.1 GBXI01003286 JAD11006.1 OUUW01000004 SPP80034.1 GAMC01010828 JAB95727.1 AK418136 BAN21351.1 KV467250 OCT56758.1 BC078024 AAH78024.1 GGMS01005170 MBY74373.1 GAKP01004074 JAC54878.1 JARO02004047 KPP69257.1 BC074301 AAH74301.1 CCAG010009633 GEFM01004607 JAP71189.1 RJVU01019149 ROL51195.1 BC154890 AAI54891.1 BC064160 AAH64160.1 U92794 BC009816 GDHF01014922 GDHF01013888 JAI37392.1 JAI38426.1 GEBF01005937 JAN97695.1 AAQR03140653 AAQR03140654 AAQR03140655 AAQR03140656 AQIA01034305 AQIA01034306 AQIA01034307 GDIP01170884 JAJ52518.1 JSUE03020790 JSUE03020791
KQ459589 KPI97471.1 KQ459896 KPJ19450.1 GAIX01004066 JAA88494.1 NWSH01002631 PCG67803.1 KZ150004 PZC75256.1 AGBW02010074 OWR49217.1 ODYU01004177 SOQ43767.1 JTDY01002064 KOB72196.1 AK403151 BAM19590.1 KQ971312 EEZ98119.2 GEZM01038259 JAV81683.1 GALX01004236 JAB64230.1 GEDC01017026 JAS20272.1 AJWK01024420 GFDF01007596 JAV06488.1 GFDF01007595 JAV06489.1 GFDF01006630 JAV07454.1 GFDF01006545 JAV07539.1 KQ435883 KOX69658.1 GFDL01008834 JAV26211.1 PYGN01001278 PSN36075.1 GDIP01221092 GDIP01213238 GDIP01137490 GDIP01130596 GDIP01130595 GDIP01127834 GDIP01113000 GDIP01061107 LRGB01002451 JAJ10164.1 KZS07500.1 GDIP01222238 GDIP01222237 GDIP01184635 GDIP01156603 GDIP01146489 GDIP01146488 GDIQ01136981 GDIP01111739 JAJ01165.1 JAL14745.1 LJIJ01000284 ODM99396.1 GDIP01251373 GDIP01221093 GDIP01140845 GDIP01140844 GDIP01137492 GDIP01137491 GDIP01130594 GDIP01113002 GDIP01113001 GDIP01074409 JAI72028.1 KQ414638 KOC66858.1 GDIP01222239 GDIP01184634 GDIP01115742 GDIP01111738 GDIP01085060 GDIP01073158 GDIP01065109 GDIP01047016 GDIP01047015 JAM56699.1 GDIQ01102850 JAL48876.1 GDIP01062603 JAM41112.1 GDIQ01208311 JAK43414.1 LBMM01002287 KMQ95026.1 GL732535 EFX84260.1 KZ288192 PBC34245.1 KB631903 ERL87019.1 APGK01042136 KB741002 ENN75729.1 GL764026 EFZ18732.1 KQ435075 KZC14414.1 KQ981685 KYN38037.1 GL441846 EFN64136.1 KQ978747 KYN28642.1 ADTU01004641 GDIQ01009391 JAN85346.1 GAPW01000816 JAC12782.1 DS232064 EDS33664.1 KQ982807 KYQ50376.1 ABLF02023169 GFXV01001525 MBW13330.1 GDIP01161692 JAJ61710.1 GDIP01025764 JAM77951.1 KQ977720 KYN00477.1 QOIP01000012 RLU16255.1 GL888207 EGI65043.1 KK107488 EZA49928.1 CH902620 EDV30921.1 CH963920 EDW78114.1 CH480822 EDW55569.1 CM002910 KMY90720.1 AE014134 AY058725 AAF53621.1 AAL13954.1 AFH03737.1 GECZ01009391 JAS60378.1 CM000158 EDW90169.1 JXLN01010490 KPM05814.1 CH379060 EAL34132.2 CH479187 EDW39782.1 CH954179 EDV54838.1 CVRI01000006 CRK88328.1 GBXI01003286 JAD11006.1 OUUW01000004 SPP80034.1 GAMC01010828 JAB95727.1 AK418136 BAN21351.1 KV467250 OCT56758.1 BC078024 AAH78024.1 GGMS01005170 MBY74373.1 GAKP01004074 JAC54878.1 JARO02004047 KPP69257.1 BC074301 AAH74301.1 CCAG010009633 GEFM01004607 JAP71189.1 RJVU01019149 ROL51195.1 BC154890 AAI54891.1 BC064160 AAH64160.1 U92794 BC009816 GDHF01014922 GDHF01013888 JAI37392.1 JAI38426.1 GEBF01005937 JAN97695.1 AAQR03140653 AAQR03140654 AAQR03140655 AAQR03140656 AQIA01034305 AQIA01034306 AQIA01034307 GDIP01170884 JAJ52518.1 JSUE03020790 JSUE03020791
Proteomes
UP000053268
UP000053240
UP000218220
UP000007151
UP000037510
UP000007266
+ More
UP000092461 UP000053105 UP000245037 UP000076858 UP000094527 UP000005203 UP000053825 UP000036403 UP000000305 UP000242457 UP000075880 UP000030742 UP000019118 UP000076502 UP000078541 UP000000311 UP000078492 UP000005205 UP000002320 UP000075809 UP000007819 UP000078542 UP000002358 UP000279307 UP000007755 UP000053097 UP000007801 UP000007798 UP000001292 UP000091820 UP000000803 UP000002282 UP000001819 UP000008744 UP000092443 UP000085678 UP000008711 UP000183832 UP000092445 UP000268350 UP000192221 UP000034805 UP000092444 UP000000589 UP000007646 UP000005225 UP000233100 UP000006718
UP000092461 UP000053105 UP000245037 UP000076858 UP000094527 UP000005203 UP000053825 UP000036403 UP000000305 UP000242457 UP000075880 UP000030742 UP000019118 UP000076502 UP000078541 UP000000311 UP000078492 UP000005205 UP000002320 UP000075809 UP000007819 UP000078542 UP000002358 UP000279307 UP000007755 UP000053097 UP000007801 UP000007798 UP000001292 UP000091820 UP000000803 UP000002282 UP000001819 UP000008744 UP000092443 UP000085678 UP000008711 UP000183832 UP000092445 UP000268350 UP000192221 UP000034805 UP000092444 UP000000589 UP000007646 UP000005225 UP000233100 UP000006718
PRIDE
Interpro
Gene 3D
ProteinModelPortal
L8B6C5
A0A1E1WUQ0
A0A345W862
A0A194PVQ6
A0A194RQ52
S4PCP7
+ More
A0A2A4J8T2 A0A2W1BRF2 A0A212F655 A0A2H1VSG9 A0A0L7LAG7 I4DNV0 D6W9R4 A0A1Y1M7I2 V5G2I9 A0A1B6D3J4 A0A1B0CR59 A0A1L8DJ45 A0A1L8DJ39 A0A1L8DM68 A0A1L8DM51 A0A0N1IT49 A0A1Q3FF99 A0A2P8XVP6 A0A0P4ZG66 A0A0P4YW50 A0A1D2N278 A0A088AUN3 A0A0P4WUM2 A0A0L7R7R0 A0A0P5Z9C2 A0A0P5RRM1 A0A0P5Y5P3 A0A0P5IT85 A0A0J7KXH9 E9G8J3 A0A2A3ER50 A0A182IWR0 U4TZH6 N6T690 E9IKT6 A0A154PSW3 A0A195FCW2 E2AQZ0 A0A195EL25 A0A158NZI4 A0A0P6I4Q2 A0A023EUW9 B0WS45 A0A151WR87 J9JPV0 A0A2H8TH21 A0A0P5DIB6 A0A0P6BRN7 A0A195CJ66 K7J4G8 A0A3L8D713 F4WL80 A0A026W1M5 B3MMH5 B4N1C4 B4I5B3 A0A1A9WM41 A0A0J9TQ58 Q9VJD1 A0A1B6GD55 B4P8R1 A0A132A4G4 Q29LB3 B4GQB0 A0A1A9Y5B2 A0A1S3JT06 B3NMG7 A0A1J1HJU0 A0A1B0A3X9 A0A0A1XJ19 A0A3B0JCE6 W8BAL3 R4WR17 A0A310TPI7 A0A1W4ULN9 Q6AZH1 A0A2S2Q9G1 A0A034WM66 A0A0N8JZG3 Q6GLZ2 A0A1B0FH60 A0A131XVY0 A0A3N0YY10 A8WGX5 Q6P373 O08795-2 A0A0K8VHR2 A0A0P6J571 G3UJW5 H0XEE9 A0A2K5WQ22 A0A0P5CQM8 F7C6C6
A0A2A4J8T2 A0A2W1BRF2 A0A212F655 A0A2H1VSG9 A0A0L7LAG7 I4DNV0 D6W9R4 A0A1Y1M7I2 V5G2I9 A0A1B6D3J4 A0A1B0CR59 A0A1L8DJ45 A0A1L8DJ39 A0A1L8DM68 A0A1L8DM51 A0A0N1IT49 A0A1Q3FF99 A0A2P8XVP6 A0A0P4ZG66 A0A0P4YW50 A0A1D2N278 A0A088AUN3 A0A0P4WUM2 A0A0L7R7R0 A0A0P5Z9C2 A0A0P5RRM1 A0A0P5Y5P3 A0A0P5IT85 A0A0J7KXH9 E9G8J3 A0A2A3ER50 A0A182IWR0 U4TZH6 N6T690 E9IKT6 A0A154PSW3 A0A195FCW2 E2AQZ0 A0A195EL25 A0A158NZI4 A0A0P6I4Q2 A0A023EUW9 B0WS45 A0A151WR87 J9JPV0 A0A2H8TH21 A0A0P5DIB6 A0A0P6BRN7 A0A195CJ66 K7J4G8 A0A3L8D713 F4WL80 A0A026W1M5 B3MMH5 B4N1C4 B4I5B3 A0A1A9WM41 A0A0J9TQ58 Q9VJD1 A0A1B6GD55 B4P8R1 A0A132A4G4 Q29LB3 B4GQB0 A0A1A9Y5B2 A0A1S3JT06 B3NMG7 A0A1J1HJU0 A0A1B0A3X9 A0A0A1XJ19 A0A3B0JCE6 W8BAL3 R4WR17 A0A310TPI7 A0A1W4ULN9 Q6AZH1 A0A2S2Q9G1 A0A034WM66 A0A0N8JZG3 Q6GLZ2 A0A1B0FH60 A0A131XVY0 A0A3N0YY10 A8WGX5 Q6P373 O08795-2 A0A0K8VHR2 A0A0P6J571 G3UJW5 H0XEE9 A0A2K5WQ22 A0A0P5CQM8 F7C6C6
PDB
5IEG
E-value=4.31322e-30,
Score=329
Ontologies
GO
PANTHER
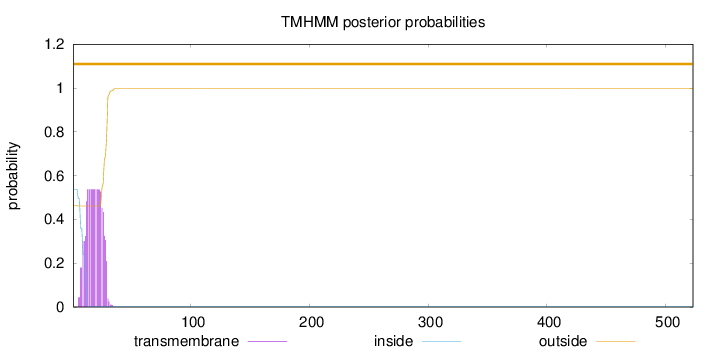
Topology
Subcellular location
Endoplasmic reticulum
SignalP
Position: 1 - 26,
Likelihood: 0.766809

Length:
523
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
10.10789
Exp number, first 60 AAs:
10.10786
Total prob of N-in:
0.53794
POSSIBLE N-term signal
sequence
outside
1 - 523
Population Genetic Test Statistics
Pi
54.585553
Theta
128.088827
Tajima's D
-1.301558
CLR
370.208842
CSRT
0.0893955302234888
Interpretation
Uncertain
