Pre Gene Modal
BGIBMGA006359
Annotation
PREDICTED:_WD_repeat-containing_protein_36_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.348 Nuclear Reliability : 1.559
Sequence
CDS
ATGAAAATGGGTCAGCACATACTGAAGGCTCCAGGCCTTATAAAAGGAGATGTTGCAACATGCCTCACTGTCACTAACTGTGGAAACTTTGTTATAATCGGTTATAGTAATGGACAAGTACACAGATTCAACATGCAATCAGGAATACACAGAGGGACTTTTGGGAATGAGAAGAAAATAGCACACAAAAGTGCCGTCAGAGGAGTTGAGACTGATTTATGTAACCAAAAGGCTATCACTGCAGGTGCAGATAATAAACTGAAATTCTGGTACTTTAAGAATTTAACTGCACCATACTCAGTTATGCGATTAGAAGAGTCAGTTTCAATGACTAAATGCCACAGAGAGAGTGCCTTGTTAGCATTGGCTAATGAGGATTTCAGTATTACTATAGTTGATATAGACACAGTGAATATAGTGAGAAAATTTGAGGGACATGCAGGAAAAATAAATGACATTGATTTTGACCACCAAAGCAGGTGGCTGATTAGTTCTTCAATGGATTCTACAATATGTACATGGGACATACCTAGTGGTGAACTTGTAGATATATTTGTAACTGAGCGACCATGTACATCCTTAAGCATGTCACCTACTGGCGATTACTTAGCCACGACGCATGTCGGTGATATAGGAGTGTTTCTGTGGGCTAACAAACTTTTGTATGAAAAGATATTCCTGAAACCAATTGACCGATATTCTGTGGATATACCTAAACTTGCTCTGCCTACATTGACACCAAAAGATCCATGTCAAAATGAAACAGAAATCATTGAAGTCTGTGAAGAACCAGAATACAAATCTCCCATCCAAATCAGTGAAGAATTAATAACTTTGTCGAATCAGCCAACGTCGCGATGGTTGAATTTATTAAACCTCGATATTGTTAAAAAACGTAATAAGCCAAAAACACCTCTGACAATACCAAAATCGGCACCTTTCTTCCTGCCCACCATACCTAGCTTAGAGCTGGAATTTGACGTAGAAAAAGATAAAGAACACAATAGTAATTCTAAGTTATTGATACCCGAATCTCTGTCGACGTTGTCAGCATTCGCCAAGAAACTAGTATCTTCTGATAGTAATGAAGACTTCAGTAAATGCGTTGAGATTTTGAAGTCTTTATCCTCGTCGGCTATTGAAGTCGAAATCAACAGTATGGCGCCTGACATGGGCGGTTCTATAGACGTAATGGTTAAATTCTTGCAAATGTTAGAGACTACATTGAACAGTAACAGAGATTTCGAATTAGCTCAATCTTATTTAAGTTTATTTTTAAAATCTCACACAAAGGTTATATCCGAGGAAAAAGAACTGCGTGATTCATTGATTTCAGTAGAAGAAGCGGCCACAGAATCATGGTCGAAATTACAAACTCAGTTGATGTATAATATCTGTATAGTTAAGGCATTAAAGGACATGTAA
Protein
MKMGQHILKAPGLIKGDVATCLTVTNCGNFVIIGYSNGQVHRFNMQSGIHRGTFGNEKKIAHKSAVRGVETDLCNQKAITAGADNKLKFWYFKNLTAPYSVMRLEESVSMTKCHRESALLALANEDFSITIVDIDTVNIVRKFEGHAGKINDIDFDHQSRWLISSSMDSTICTWDIPSGELVDIFVTERPCTSLSMSPTGDYLATTHVGDIGVFLWANKLLYEKIFLKPIDRYSVDIPKLALPTLTPKDPCQNETEIIEVCEEPEYKSPIQISEELITLSNQPTSRWLNLLNLDIVKKRNKPKTPLTIPKSAPFFLPTIPSLELEFDVEKDKEHNSNSKLLIPESLSTLSAFAKKLVSSDSNEDFSKCVEILKSLSSSAIEVEINSMAPDMGGSIDVMVKFLQMLETTLNSNRDFELAQSYLSLFLKSHTKVISEEKELRDSLISVEEAATESWSKLQTQLMYNICIVKALKDM
Summary
Uniprot
H9JA14
A0A2H1VSE6
A0A2W1BK48
A0A2A4J6U5
A0A194RNN4
A0A194PWS8
+ More
A0A212F654 A0A2J7Q9L4 A0A0C9PWV5 A0A067RF02 A0A0L0CH26 A0A0M8ZXP0 A0A2P8XU64 T1PI57 A0A1I8MSS4 A0A195F115 D6WA27 A0A0L7QLK8 A0A195DXM2 V5HB96 A0A151ILD5 A0A088AG54 A0A2A3EFB5 A0A151X8B7 A0A0M4EI30 A0A232FHL5 K7JGZ9 A0A158NP15 E2BZ01 A0A1B6CU23 A0A1Y1JXJ9 A0A154NWL2 F4W8X4 A0A195BYD7 A0A131XG89 M7BPT4 Q16W76 A0A1S4FLX1 A0A1E1XJ16 A0A026VX66 A0A1W4XNP5 A0A1E1XB67 A0A3L8DHG1 E9IED0 A0A131YWR2 U4UPS4 N6TDF3 B3LX80 A0A093FMM8 A0A0B6Z6J4 A0A147BQV4 A0A094L2W2 B4M6B4 A0A091KRF5 A0A1Q3EZB5 A0A091G160 A0A1U7RZV2 A0A0J7KNA4 A0A0A0B0G0 A0A310SV31 A0A087RBK8 A0A182H6A8 A0A093I667 A0A336LZ66 A0A224ZBY7 A0A151PI76 A0A1S3H3P4 L7M9G2 A0A091JH06 A0A091J9Y6 B4JI72 A0A2I0UMN5 A0A034VP19 A0A336L6A0 A0A2P4TBY1 A0A091WI26 A0A093P012 Q960D6 A0A1B6JF22 A0A2K5S518 A0A2M4AHX6 A0A093PPW6 H0YRW1 A0A218UUH1 A0A2R2MKY5 Q9VFV8 A0A093CM77 A0A0K8UBZ4 A0A1W4V3D6 A0A094N2X5 A0A093QUJ4 A0A091UME9 A0A2I0M8X8 A0A2M4BCR9 E2ABT5 A0A2M4BCP1 A0A1B6FF83 R0K351 U3IZN1
A0A212F654 A0A2J7Q9L4 A0A0C9PWV5 A0A067RF02 A0A0L0CH26 A0A0M8ZXP0 A0A2P8XU64 T1PI57 A0A1I8MSS4 A0A195F115 D6WA27 A0A0L7QLK8 A0A195DXM2 V5HB96 A0A151ILD5 A0A088AG54 A0A2A3EFB5 A0A151X8B7 A0A0M4EI30 A0A232FHL5 K7JGZ9 A0A158NP15 E2BZ01 A0A1B6CU23 A0A1Y1JXJ9 A0A154NWL2 F4W8X4 A0A195BYD7 A0A131XG89 M7BPT4 Q16W76 A0A1S4FLX1 A0A1E1XJ16 A0A026VX66 A0A1W4XNP5 A0A1E1XB67 A0A3L8DHG1 E9IED0 A0A131YWR2 U4UPS4 N6TDF3 B3LX80 A0A093FMM8 A0A0B6Z6J4 A0A147BQV4 A0A094L2W2 B4M6B4 A0A091KRF5 A0A1Q3EZB5 A0A091G160 A0A1U7RZV2 A0A0J7KNA4 A0A0A0B0G0 A0A310SV31 A0A087RBK8 A0A182H6A8 A0A093I667 A0A336LZ66 A0A224ZBY7 A0A151PI76 A0A1S3H3P4 L7M9G2 A0A091JH06 A0A091J9Y6 B4JI72 A0A2I0UMN5 A0A034VP19 A0A336L6A0 A0A2P4TBY1 A0A091WI26 A0A093P012 Q960D6 A0A1B6JF22 A0A2K5S518 A0A2M4AHX6 A0A093PPW6 H0YRW1 A0A218UUH1 A0A2R2MKY5 Q9VFV8 A0A093CM77 A0A0K8UBZ4 A0A1W4V3D6 A0A094N2X5 A0A093QUJ4 A0A091UME9 A0A2I0M8X8 A0A2M4BCR9 E2ABT5 A0A2M4BCP1 A0A1B6FF83 R0K351 U3IZN1
Pubmed
19121390
28756777
26354079
22118469
24845553
26108605
+ More
29403074 25315136 18362917 19820115 25765539 28648823 20075255 21347285 20798317 28004739 21719571 28049606 23624526 17510324 29209593 24508170 28503490 30249741 21282665 26830274 23537049 17994087 29652888 26483478 28797301 22293439 25576852 25348373 20360741 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23371554 23749191
29403074 25315136 18362917 19820115 25765539 28648823 20075255 21347285 20798317 28004739 21719571 28049606 23624526 17510324 29209593 24508170 28503490 30249741 21282665 26830274 23537049 17994087 29652888 26483478 28797301 22293439 25576852 25348373 20360741 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23371554 23749191
EMBL
BABH01009394
ODYU01004177
SOQ43765.1
KZ150004
PZC75258.1
NWSH01002631
+ More
PCG67805.1 KQ459896 KPJ19448.1 KQ459589 KPI97473.1 AGBW02010074 OWR49220.1 NEVH01016348 PNF25272.1 GBYB01005988 JAG75755.1 KK852510 KDR22347.1 JRES01000409 KNC31502.1 KQ435801 KOX73225.1 PYGN01001339 PSN35551.1 KA648451 AFP63080.1 KQ981891 KYN33794.1 KQ971312 EEZ98071.1 KQ414914 KOC59513.1 KQ980167 KYN17364.1 GANP01012296 JAB72172.1 KQ977120 KYN05563.1 KZ288262 PBC30427.1 KQ982422 KYQ56621.1 CP012526 ALC46221.1 NNAY01000180 OXU30241.1 ADTU01022032 GL451538 EFN79073.1 GEDC01020259 JAS17039.1 GEZM01097920 JAV54042.1 KQ434773 KZC03983.1 GL887974 EGI69333.1 KQ976394 KYM93320.1 GEFH01002984 JAP65597.1 KB515362 EMP39921.1 CH477573 EAT38834.1 GFAA01004134 JAT99300.1 KK107648 EZA48352.1 GFAC01002710 JAT96478.1 QOIP01000008 RLU19781.1 GL762576 EFZ21087.1 GEDV01005469 JAP83088.1 KB632309 ERL92025.1 APGK01042225 KB741002 ENN75768.1 CH902617 EDV41680.1 KK634978 KFV57895.1 HACG01017112 CEK63977.1 GEGO01002257 JAR93147.1 KL274746 KFZ65698.1 CH940652 EDW59190.1 KK751966 KFP42078.1 GFDL01014467 JAV20578.1 KL447644 KFO75723.1 LBMM01005134 KMQ91792.1 KL884777 KGM00285.1 KQ760166 OAD61660.1 KL226279 KFM10862.1 JXUM01113766 KQ565724 KXJ70732.1 KL206890 KFV86692.1 UFQT01000244 SSX22311.1 GFPF01013366 MAA24512.1 AKHW03000179 KYO48797.1 GACK01005246 JAA59788.1 KK500856 KFP10961.1 KL218793 KFP08521.1 CH916369 EDV92953.1 KZ505682 PKU47283.1 GAKP01015090 JAC43862.1 UFQS01001136 UFQT01001136 SSX09170.1 SSX29080.1 PPHD01002783 POI33864.1 KK733913 KFR01236.1 KL225272 KFW70373.1 AY052114 AAK93538.1 GECU01009968 JAS97738.1 GGFK01007074 MBW40395.1 KL670372 KFW78888.1 ABQF01032508 ABQF01032509 ABQF01032510 MUZQ01000124 OWK57395.1 AE014297 BT016079 AAF54941.1 AAV36964.1 KL462590 KFV14044.1 GDHF01028206 JAI24108.1 KL354261 KFZ61046.1 KL424985 KFW90040.1 KL409831 KFQ92119.1 AKCR02000029 PKK26133.1 GGFJ01001676 MBW50817.1 GL438356 EFN69112.1 GGFJ01001678 MBW50819.1 GECZ01020908 JAS48861.1 KB742782 EOB04466.1 ADON01043173 ADON01043174 ADON01043175 ADON01043176 ADON01043177 ADON01043178 ADON01043179 ADON01043180 ADON01043181 ADON01043182
PCG67805.1 KQ459896 KPJ19448.1 KQ459589 KPI97473.1 AGBW02010074 OWR49220.1 NEVH01016348 PNF25272.1 GBYB01005988 JAG75755.1 KK852510 KDR22347.1 JRES01000409 KNC31502.1 KQ435801 KOX73225.1 PYGN01001339 PSN35551.1 KA648451 AFP63080.1 KQ981891 KYN33794.1 KQ971312 EEZ98071.1 KQ414914 KOC59513.1 KQ980167 KYN17364.1 GANP01012296 JAB72172.1 KQ977120 KYN05563.1 KZ288262 PBC30427.1 KQ982422 KYQ56621.1 CP012526 ALC46221.1 NNAY01000180 OXU30241.1 ADTU01022032 GL451538 EFN79073.1 GEDC01020259 JAS17039.1 GEZM01097920 JAV54042.1 KQ434773 KZC03983.1 GL887974 EGI69333.1 KQ976394 KYM93320.1 GEFH01002984 JAP65597.1 KB515362 EMP39921.1 CH477573 EAT38834.1 GFAA01004134 JAT99300.1 KK107648 EZA48352.1 GFAC01002710 JAT96478.1 QOIP01000008 RLU19781.1 GL762576 EFZ21087.1 GEDV01005469 JAP83088.1 KB632309 ERL92025.1 APGK01042225 KB741002 ENN75768.1 CH902617 EDV41680.1 KK634978 KFV57895.1 HACG01017112 CEK63977.1 GEGO01002257 JAR93147.1 KL274746 KFZ65698.1 CH940652 EDW59190.1 KK751966 KFP42078.1 GFDL01014467 JAV20578.1 KL447644 KFO75723.1 LBMM01005134 KMQ91792.1 KL884777 KGM00285.1 KQ760166 OAD61660.1 KL226279 KFM10862.1 JXUM01113766 KQ565724 KXJ70732.1 KL206890 KFV86692.1 UFQT01000244 SSX22311.1 GFPF01013366 MAA24512.1 AKHW03000179 KYO48797.1 GACK01005246 JAA59788.1 KK500856 KFP10961.1 KL218793 KFP08521.1 CH916369 EDV92953.1 KZ505682 PKU47283.1 GAKP01015090 JAC43862.1 UFQS01001136 UFQT01001136 SSX09170.1 SSX29080.1 PPHD01002783 POI33864.1 KK733913 KFR01236.1 KL225272 KFW70373.1 AY052114 AAK93538.1 GECU01009968 JAS97738.1 GGFK01007074 MBW40395.1 KL670372 KFW78888.1 ABQF01032508 ABQF01032509 ABQF01032510 MUZQ01000124 OWK57395.1 AE014297 BT016079 AAF54941.1 AAV36964.1 KL462590 KFV14044.1 GDHF01028206 JAI24108.1 KL354261 KFZ61046.1 KL424985 KFW90040.1 KL409831 KFQ92119.1 AKCR02000029 PKK26133.1 GGFJ01001676 MBW50817.1 GL438356 EFN69112.1 GGFJ01001678 MBW50819.1 GECZ01020908 JAS48861.1 KB742782 EOB04466.1 ADON01043173 ADON01043174 ADON01043175 ADON01043176 ADON01043177 ADON01043178 ADON01043179 ADON01043180 ADON01043181 ADON01043182
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000235965
+ More
UP000027135 UP000037069 UP000053105 UP000245037 UP000095301 UP000078541 UP000007266 UP000053825 UP000078492 UP000078542 UP000005203 UP000242457 UP000075809 UP000092553 UP000215335 UP000002358 UP000005205 UP000008237 UP000076502 UP000007755 UP000078540 UP000031443 UP000008820 UP000053097 UP000192223 UP000279307 UP000030742 UP000019118 UP000007801 UP000008792 UP000053760 UP000189705 UP000036403 UP000053858 UP000053286 UP000069940 UP000249989 UP000053584 UP000050525 UP000085678 UP000053119 UP000054308 UP000001070 UP000053605 UP000054081 UP000233040 UP000053258 UP000007754 UP000197619 UP000000803 UP000192221 UP000053283 UP000053872 UP000000311 UP000016666
UP000027135 UP000037069 UP000053105 UP000245037 UP000095301 UP000078541 UP000007266 UP000053825 UP000078492 UP000078542 UP000005203 UP000242457 UP000075809 UP000092553 UP000215335 UP000002358 UP000005205 UP000008237 UP000076502 UP000007755 UP000078540 UP000031443 UP000008820 UP000053097 UP000192223 UP000279307 UP000030742 UP000019118 UP000007801 UP000008792 UP000053760 UP000189705 UP000036403 UP000053858 UP000053286 UP000069940 UP000249989 UP000053584 UP000050525 UP000085678 UP000053119 UP000054308 UP000001070 UP000053605 UP000054081 UP000233040 UP000053258 UP000007754 UP000197619 UP000000803 UP000192221 UP000053283 UP000053872 UP000000311 UP000016666
Interpro
IPR019775
WD40_repeat_CS
+ More
IPR036322 WD40_repeat_dom_sf
IPR007319 SSU_processome_Utp21
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR024977 Apc4_WD40_dom
IPR011047 Quinoprotein_ADH-like_supfam
IPR011043 Gal_Oxase/kelch_b-propeller
IPR020472 G-protein_beta_WD-40_rep
IPR002372 PQQ_repeat
IPR020719 RNA3'_term_phos_cycl-like_CS
IPR036322 WD40_repeat_dom_sf
IPR007319 SSU_processome_Utp21
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR024977 Apc4_WD40_dom
IPR011047 Quinoprotein_ADH-like_supfam
IPR011043 Gal_Oxase/kelch_b-propeller
IPR020472 G-protein_beta_WD-40_rep
IPR002372 PQQ_repeat
IPR020719 RNA3'_term_phos_cycl-like_CS
Gene 3D
ProteinModelPortal
H9JA14
A0A2H1VSE6
A0A2W1BK48
A0A2A4J6U5
A0A194RNN4
A0A194PWS8
+ More
A0A212F654 A0A2J7Q9L4 A0A0C9PWV5 A0A067RF02 A0A0L0CH26 A0A0M8ZXP0 A0A2P8XU64 T1PI57 A0A1I8MSS4 A0A195F115 D6WA27 A0A0L7QLK8 A0A195DXM2 V5HB96 A0A151ILD5 A0A088AG54 A0A2A3EFB5 A0A151X8B7 A0A0M4EI30 A0A232FHL5 K7JGZ9 A0A158NP15 E2BZ01 A0A1B6CU23 A0A1Y1JXJ9 A0A154NWL2 F4W8X4 A0A195BYD7 A0A131XG89 M7BPT4 Q16W76 A0A1S4FLX1 A0A1E1XJ16 A0A026VX66 A0A1W4XNP5 A0A1E1XB67 A0A3L8DHG1 E9IED0 A0A131YWR2 U4UPS4 N6TDF3 B3LX80 A0A093FMM8 A0A0B6Z6J4 A0A147BQV4 A0A094L2W2 B4M6B4 A0A091KRF5 A0A1Q3EZB5 A0A091G160 A0A1U7RZV2 A0A0J7KNA4 A0A0A0B0G0 A0A310SV31 A0A087RBK8 A0A182H6A8 A0A093I667 A0A336LZ66 A0A224ZBY7 A0A151PI76 A0A1S3H3P4 L7M9G2 A0A091JH06 A0A091J9Y6 B4JI72 A0A2I0UMN5 A0A034VP19 A0A336L6A0 A0A2P4TBY1 A0A091WI26 A0A093P012 Q960D6 A0A1B6JF22 A0A2K5S518 A0A2M4AHX6 A0A093PPW6 H0YRW1 A0A218UUH1 A0A2R2MKY5 Q9VFV8 A0A093CM77 A0A0K8UBZ4 A0A1W4V3D6 A0A094N2X5 A0A093QUJ4 A0A091UME9 A0A2I0M8X8 A0A2M4BCR9 E2ABT5 A0A2M4BCP1 A0A1B6FF83 R0K351 U3IZN1
A0A212F654 A0A2J7Q9L4 A0A0C9PWV5 A0A067RF02 A0A0L0CH26 A0A0M8ZXP0 A0A2P8XU64 T1PI57 A0A1I8MSS4 A0A195F115 D6WA27 A0A0L7QLK8 A0A195DXM2 V5HB96 A0A151ILD5 A0A088AG54 A0A2A3EFB5 A0A151X8B7 A0A0M4EI30 A0A232FHL5 K7JGZ9 A0A158NP15 E2BZ01 A0A1B6CU23 A0A1Y1JXJ9 A0A154NWL2 F4W8X4 A0A195BYD7 A0A131XG89 M7BPT4 Q16W76 A0A1S4FLX1 A0A1E1XJ16 A0A026VX66 A0A1W4XNP5 A0A1E1XB67 A0A3L8DHG1 E9IED0 A0A131YWR2 U4UPS4 N6TDF3 B3LX80 A0A093FMM8 A0A0B6Z6J4 A0A147BQV4 A0A094L2W2 B4M6B4 A0A091KRF5 A0A1Q3EZB5 A0A091G160 A0A1U7RZV2 A0A0J7KNA4 A0A0A0B0G0 A0A310SV31 A0A087RBK8 A0A182H6A8 A0A093I667 A0A336LZ66 A0A224ZBY7 A0A151PI76 A0A1S3H3P4 L7M9G2 A0A091JH06 A0A091J9Y6 B4JI72 A0A2I0UMN5 A0A034VP19 A0A336L6A0 A0A2P4TBY1 A0A091WI26 A0A093P012 Q960D6 A0A1B6JF22 A0A2K5S518 A0A2M4AHX6 A0A093PPW6 H0YRW1 A0A218UUH1 A0A2R2MKY5 Q9VFV8 A0A093CM77 A0A0K8UBZ4 A0A1W4V3D6 A0A094N2X5 A0A093QUJ4 A0A091UME9 A0A2I0M8X8 A0A2M4BCR9 E2ABT5 A0A2M4BCP1 A0A1B6FF83 R0K351 U3IZN1
PDB
5OQL
E-value=1.28717e-55,
Score=548
Ontologies
GO
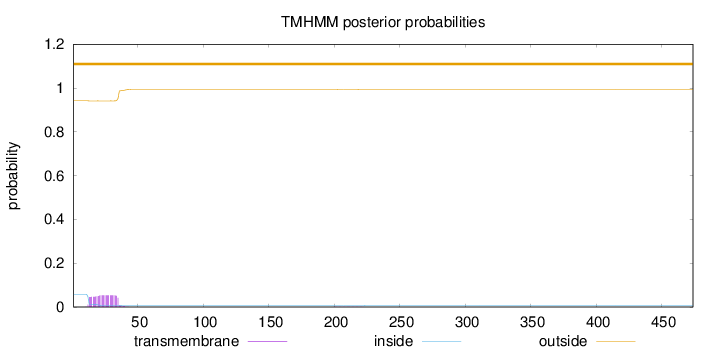
Topology
Length:
474
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.20816
Exp number, first 60 AAs:
1.2009
Total prob of N-in:
0.05839
outside
1 - 474
Population Genetic Test Statistics
Pi
185.638403
Theta
167.909422
Tajima's D
0.39215
CLR
0.67331
CSRT
0.487125643717814
Interpretation
Uncertain
