Gene
KWMTBOMO03467
Annotation
Sugar_transporter_[Operophtera_brumata]
Location in the cell
PlasmaMembrane Reliability : 4.968
Sequence
CDS
ATGGATTCTTGTGCTGACGAAGCAGTTGATTTGAATGAAAGTGGTGAAGCAGACGATAAGAATAGCGTTACAACAAAGAGATATCTATGGCGACAAATTTTTGTTTCAAGTGGAGTTTGGTCGATTTACTTTGTTCTTGGTTTGGGCTTCGGAGCTCCAACTGTAATGATCCCACAGATCAGAAGAGAAGCTAACTCAACAGATGCAGTTACCGAGAGCATGGCTTCATGGCTAACTTCCGTGCACGGCTACTCTGCACTTCCATGGGTATTCGTGATACCCGTTTTGACAAGATACATCGGAAGAAAAATACCTTTCACCATAATATGTATAGTGACTTTAGTTTCGTTCCTGTTATTTTATTTCAGTACAAATACAACGCATTTATTGATCAGTGCGATAATGCAAGGGATGCTTCTAGCAAGCAACATGACTTTGTTGGTTGTTATTGTAACGGAATACTCCTCCCCGAAATATCGAGGGGTATTCATGATATTCGAATCTACAGTTTTCTTTTGGGGAGTATGGGTCGCTAATGCGACGGGAGCCTTTTCCCACTGGAAAAACATCGGGTACATCGCCTTTGTTTGTTCTCTGTACCCTTTGACCGTCGTATTTTGGCCCGAATCGCCTTACTGGTTGGCGATGAAGGGCCGGTTCGAGGAATGTGCCGCTTCACACCACTGGCTGAAAGGGTACGATAAAGATTCCCAATGTGAACTCGAGACGTTGATTGATTCTCAGAAACAGTATATTAAGATGTGTGCGGAGAATAAACGAAATGTTGCAGGGAATAGAGTGAAGTTTATTTATGAAACTATAAGCACAAAAGCGTTTTACATGCCAATGTTTATATCTGTTGTAGTAATGTCAATGTATCATTTCTCTGGAAAATTAGTATGCACGATGTATGCGATGGATTTAATTAATCAAATAACTGGCAGCAACATTACCTCAAGTTATGGCATGCTGATTATGGACATCATTACAATAGTAGGTATGCAGCTCGGATGTATTTTATCAAAGTTTTTGAAAAGGCGTACATTGTTGTTGACTTCGGCCACTTCGGGGATTTTCTTTCTCTACGTCATATCTGTATATTTGTATCTAGTTAAGTATTCGGTTATTGCCGAGAATAGATATTTGTCAATATTGTTTCTGACATTGTTTTCCGTGTCAATAAGTCTCGGTCCCATGATAATGCCGTCCACTATATTTGGAGAGTTAATATGTTTAAGGTACCAAACGCCTTGTTTACTTTTATTAACATTGTTTTCTGAATTTTTAATGGCAACTATTTTGAAAACATCTCCGTATATATTTCGTACCCTAACTTTACCCGGTGCTTTCCTGTTTTACGGGCTATCAGCATCTTTTTTCGCATTACTTGTTTACAAATATTTACCGGAAACTAAAGATAAGACGTTATTTGAGATTGAGAATTATTTCAAAGATCCCAATCAGAGACAAAACGATGTTGAGAGGGTTTTGATAAAGCATAGGTAA
Protein
MDSCADEAVDLNESGEADDKNSVTTKRYLWRQIFVSSGVWSIYFVLGLGFGAPTVMIPQIRREANSTDAVTESMASWLTSVHGYSALPWVFVIPVLTRYIGRKIPFTIICIVTLVSFLLFYFSTNTTHLLISAIMQGMLLASNMTLLVVIVTEYSSPKYRGVFMIFESTVFFWGVWVANATGAFSHWKNIGYIAFVCSLYPLTVVFWPESPYWLAMKGRFEECAASHHWLKGYDKDSQCELETLIDSQKQYIKMCAENKRNVAGNRVKFIYETISTKAFYMPMFISVVVMSMYHFSGKLVCTMYAMDLINQITGSNITSSYGMLIMDIITIVGMQLGCILSKFLKRRTLLLTSATSGIFFLYVISVYLYLVKYSVIAENRYLSILFLTLFSVSISLGPMIMPSTIFGELICLRYQTPCLLLLTLFSEFLMATILKTSPYIFRTLTLPGAFLFYGLSASFFALLVYKYLPETKDKTLFEIENYFKDPNQRQNDVERVLIKHR
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
A0A2A4JBS9
A0A2W1BJK8
A0A2W1BJN4
A0A2H1VQR0
A0A0L7KW08
A0A2W1BLU7
+ More
A0A2H1VQR9 A0A2W1BR72 H9JAI4 A0A0L7KPS0 H9IWZ7 A0A2H1X3V1 H9JFD5 A0A2H1X3Y1 A0A2W1BQ15 A0A2A4K3Q9 A0A2H1VYW0 A0A2H1VYX5 A0A2A4K454 A0A212EJQ6 A0A0L7LW97 A0A194Q2H3 A0A0L7LSQ8 A0A0L7KRG0 A0A0L7LBU2 A0A194R9C4 A0A212ETB8 A0A0N1IQT4 A0A0L7LC11 A0A2H1WKZ9 A0A0L7KWB8 A0A2W1B9E8 A0A2A4KA42 A0A194QKY0 A0A2H1WK46 A0A0N1I9K7 A0A212F9C9 A0A2A4KB29 A0A194QJ23 A0A0L7KVZ8 H9JMX8 A0A023PNK9 A0A0N1IHN6 A0A194QJI7 A0A2A4JNG3 A0A0L7KJ46 A0A212F161 A0A194QPY2 A0A2A4JTG8 A0A0L7LCK3 A0A212FJ74 A0A0N0PEP7 A0A2H1W2F0 A0A0N0PEE0 A0A0N1IPS2 A0A2H1V5D8 A0A194QHU0 A0A212ENJ2 A0A2H1WVP2 A0A0L7LFR7 A0A336M7L2 A0A0N1IG19 A0A2A4JUU3 A0A2A4KAY2 A0A2H1WX63 A0A2W1BF54 A0A194QJ80 H9J017 H9J4S1 A0A2W1BG84 A0A2W1BN20 A0A336MJD0 A0A0L7KUI7 A0A0L7LHM6 A0A194QJ30 A0A2W1BD78 H9J4X7 A0A1L8DFA5 A0A0L7KLX5 A0A0N0PAH4 H9J539 A0A2H1W936 H9IV67 A0A0L7KWI6 A0A1B0CWH0 A0A1L8DF47 A0A2A4IVM9 A0A3G1T1H2 H9J4T7 A0A2H1WFL6 A0A2A4JT91 A0A2A4JKJ9 H9J4U8 A0A336MDU0 A0A139WGJ0
A0A2H1VQR9 A0A2W1BR72 H9JAI4 A0A0L7KPS0 H9IWZ7 A0A2H1X3V1 H9JFD5 A0A2H1X3Y1 A0A2W1BQ15 A0A2A4K3Q9 A0A2H1VYW0 A0A2H1VYX5 A0A2A4K454 A0A212EJQ6 A0A0L7LW97 A0A194Q2H3 A0A0L7LSQ8 A0A0L7KRG0 A0A0L7LBU2 A0A194R9C4 A0A212ETB8 A0A0N1IQT4 A0A0L7LC11 A0A2H1WKZ9 A0A0L7KWB8 A0A2W1B9E8 A0A2A4KA42 A0A194QKY0 A0A2H1WK46 A0A0N1I9K7 A0A212F9C9 A0A2A4KB29 A0A194QJ23 A0A0L7KVZ8 H9JMX8 A0A023PNK9 A0A0N1IHN6 A0A194QJI7 A0A2A4JNG3 A0A0L7KJ46 A0A212F161 A0A194QPY2 A0A2A4JTG8 A0A0L7LCK3 A0A212FJ74 A0A0N0PEP7 A0A2H1W2F0 A0A0N0PEE0 A0A0N1IPS2 A0A2H1V5D8 A0A194QHU0 A0A212ENJ2 A0A2H1WVP2 A0A0L7LFR7 A0A336M7L2 A0A0N1IG19 A0A2A4JUU3 A0A2A4KAY2 A0A2H1WX63 A0A2W1BF54 A0A194QJ80 H9J017 H9J4S1 A0A2W1BG84 A0A2W1BN20 A0A336MJD0 A0A0L7KUI7 A0A0L7LHM6 A0A194QJ30 A0A2W1BD78 H9J4X7 A0A1L8DFA5 A0A0L7KLX5 A0A0N0PAH4 H9J539 A0A2H1W936 H9IV67 A0A0L7KWI6 A0A1B0CWH0 A0A1L8DF47 A0A2A4IVM9 A0A3G1T1H2 H9J4T7 A0A2H1WFL6 A0A2A4JT91 A0A2A4JKJ9 H9J4U8 A0A336MDU0 A0A139WGJ0
EMBL
NWSH01001973
PCG69525.1
KZ150004
PZC75262.1
PZC75259.1
ODYU01003864
+ More
SOQ43159.1 JTDY01005057 KOB67418.1 PZC75261.1 SOQ43160.1 PZC75260.1 BABH01009421 JTDY01007576 KOB65105.1 BABH01021167 ODYU01013263 SOQ59969.1 BABH01038306 SOQ59968.1 KZ149985 PZC75704.1 NWSH01000180 PCG78689.1 ODYU01005293 SOQ46008.1 ODYU01005292 SOQ46007.1 PCG78688.1 AGBW02014405 OWR41733.1 JTDY01000013 KOB79431.1 KQ459589 KPI97605.1 JTDY01000181 KOB78419.1 JTDY01006862 KOB65636.1 JTDY01001793 KOB72864.1 KQ460500 KPJ14232.1 AGBW02012599 OWR44736.1 LADJ01036943 KPJ21300.1 JTDY01001770 KOB72935.1 ODYU01009373 SOQ53753.1 JTDY01005064 KOB67410.1 KZ150249 PZC71838.1 NWSH01000008 PCG80949.1 KQ458671 KPJ05560.1 ODYU01009098 SOQ53272.1 KQ460077 KPJ17880.1 AGBW02009614 OWR50323.1 PCG80950.1 KPJ05558.1 KOB67408.1 BABH01028994 KJ439227 KZ150491 AHX25878.1 PZC70688.1 KPJ17887.1 KPJ05554.1 NWSH01001042 PCG72950.1 JTDY01009531 KOB63358.1 AGBW02010956 OWR47467.1 KPJ05561.1 NWSH01000704 PCG74682.1 KOB72931.1 AGBW02008317 OWR53759.1 KQ459890 KPJ19577.1 ODYU01005893 SOQ47251.1 KPJ19576.1 KPJ17881.1 ODYU01000544 SOQ35484.1 KQ458981 KPJ04505.1 AGBW02013662 OWR43027.1 ODYU01011356 SOQ57052.1 JTDY01001315 KOB74225.1 UFQT01000224 SSX22028.1 KPJ17891.1 NWSH01000578 PCG75536.1 PCG80953.1 ODYU01011726 SOQ57638.1 PZC71837.1 KPJ05572.1 BABH01018801 BABH01035527 BABH01035528 BABH01035529 PZC71836.1 KZ150067 PZC74120.1 UFQT01001316 SSX30005.1 JTDY01005517 KOB66923.1 JTDY01001035 KOB75068.1 KPJ05563.1 PZC71835.1 BABH01025717 GFDF01009027 JAV05057.1 JTDY01009224 KOB63914.1 KQ458842 KPJ04897.1 BABH01038877 BABH01038878 BABH01038879 BABH01038880 BABH01038881 ODYU01007122 SOQ49615.1 BABH01000587 KOB67409.1 AJWK01032461 GFDF01009026 JAV05058.1 NWSH01006268 PCG63546.1 MG992391 AXY94829.1 BABH01035469 ODYU01008354 SOQ51875.1 PCG74683.1 NWSH01001160 PCG72306.1 BABH01035543 UFQT01001016 SSX28504.1 KQ971345 KYB27009.1
SOQ43159.1 JTDY01005057 KOB67418.1 PZC75261.1 SOQ43160.1 PZC75260.1 BABH01009421 JTDY01007576 KOB65105.1 BABH01021167 ODYU01013263 SOQ59969.1 BABH01038306 SOQ59968.1 KZ149985 PZC75704.1 NWSH01000180 PCG78689.1 ODYU01005293 SOQ46008.1 ODYU01005292 SOQ46007.1 PCG78688.1 AGBW02014405 OWR41733.1 JTDY01000013 KOB79431.1 KQ459589 KPI97605.1 JTDY01000181 KOB78419.1 JTDY01006862 KOB65636.1 JTDY01001793 KOB72864.1 KQ460500 KPJ14232.1 AGBW02012599 OWR44736.1 LADJ01036943 KPJ21300.1 JTDY01001770 KOB72935.1 ODYU01009373 SOQ53753.1 JTDY01005064 KOB67410.1 KZ150249 PZC71838.1 NWSH01000008 PCG80949.1 KQ458671 KPJ05560.1 ODYU01009098 SOQ53272.1 KQ460077 KPJ17880.1 AGBW02009614 OWR50323.1 PCG80950.1 KPJ05558.1 KOB67408.1 BABH01028994 KJ439227 KZ150491 AHX25878.1 PZC70688.1 KPJ17887.1 KPJ05554.1 NWSH01001042 PCG72950.1 JTDY01009531 KOB63358.1 AGBW02010956 OWR47467.1 KPJ05561.1 NWSH01000704 PCG74682.1 KOB72931.1 AGBW02008317 OWR53759.1 KQ459890 KPJ19577.1 ODYU01005893 SOQ47251.1 KPJ19576.1 KPJ17881.1 ODYU01000544 SOQ35484.1 KQ458981 KPJ04505.1 AGBW02013662 OWR43027.1 ODYU01011356 SOQ57052.1 JTDY01001315 KOB74225.1 UFQT01000224 SSX22028.1 KPJ17891.1 NWSH01000578 PCG75536.1 PCG80953.1 ODYU01011726 SOQ57638.1 PZC71837.1 KPJ05572.1 BABH01018801 BABH01035527 BABH01035528 BABH01035529 PZC71836.1 KZ150067 PZC74120.1 UFQT01001316 SSX30005.1 JTDY01005517 KOB66923.1 JTDY01001035 KOB75068.1 KPJ05563.1 PZC71835.1 BABH01025717 GFDF01009027 JAV05057.1 JTDY01009224 KOB63914.1 KQ458842 KPJ04897.1 BABH01038877 BABH01038878 BABH01038879 BABH01038880 BABH01038881 ODYU01007122 SOQ49615.1 BABH01000587 KOB67409.1 AJWK01032461 GFDF01009026 JAV05058.1 NWSH01006268 PCG63546.1 MG992391 AXY94829.1 BABH01035469 ODYU01008354 SOQ51875.1 PCG74683.1 NWSH01001160 PCG72306.1 BABH01035543 UFQT01001016 SSX28504.1 KQ971345 KYB27009.1
Proteomes
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A2A4JBS9
A0A2W1BJK8
A0A2W1BJN4
A0A2H1VQR0
A0A0L7KW08
A0A2W1BLU7
+ More
A0A2H1VQR9 A0A2W1BR72 H9JAI4 A0A0L7KPS0 H9IWZ7 A0A2H1X3V1 H9JFD5 A0A2H1X3Y1 A0A2W1BQ15 A0A2A4K3Q9 A0A2H1VYW0 A0A2H1VYX5 A0A2A4K454 A0A212EJQ6 A0A0L7LW97 A0A194Q2H3 A0A0L7LSQ8 A0A0L7KRG0 A0A0L7LBU2 A0A194R9C4 A0A212ETB8 A0A0N1IQT4 A0A0L7LC11 A0A2H1WKZ9 A0A0L7KWB8 A0A2W1B9E8 A0A2A4KA42 A0A194QKY0 A0A2H1WK46 A0A0N1I9K7 A0A212F9C9 A0A2A4KB29 A0A194QJ23 A0A0L7KVZ8 H9JMX8 A0A023PNK9 A0A0N1IHN6 A0A194QJI7 A0A2A4JNG3 A0A0L7KJ46 A0A212F161 A0A194QPY2 A0A2A4JTG8 A0A0L7LCK3 A0A212FJ74 A0A0N0PEP7 A0A2H1W2F0 A0A0N0PEE0 A0A0N1IPS2 A0A2H1V5D8 A0A194QHU0 A0A212ENJ2 A0A2H1WVP2 A0A0L7LFR7 A0A336M7L2 A0A0N1IG19 A0A2A4JUU3 A0A2A4KAY2 A0A2H1WX63 A0A2W1BF54 A0A194QJ80 H9J017 H9J4S1 A0A2W1BG84 A0A2W1BN20 A0A336MJD0 A0A0L7KUI7 A0A0L7LHM6 A0A194QJ30 A0A2W1BD78 H9J4X7 A0A1L8DFA5 A0A0L7KLX5 A0A0N0PAH4 H9J539 A0A2H1W936 H9IV67 A0A0L7KWI6 A0A1B0CWH0 A0A1L8DF47 A0A2A4IVM9 A0A3G1T1H2 H9J4T7 A0A2H1WFL6 A0A2A4JT91 A0A2A4JKJ9 H9J4U8 A0A336MDU0 A0A139WGJ0
A0A2H1VQR9 A0A2W1BR72 H9JAI4 A0A0L7KPS0 H9IWZ7 A0A2H1X3V1 H9JFD5 A0A2H1X3Y1 A0A2W1BQ15 A0A2A4K3Q9 A0A2H1VYW0 A0A2H1VYX5 A0A2A4K454 A0A212EJQ6 A0A0L7LW97 A0A194Q2H3 A0A0L7LSQ8 A0A0L7KRG0 A0A0L7LBU2 A0A194R9C4 A0A212ETB8 A0A0N1IQT4 A0A0L7LC11 A0A2H1WKZ9 A0A0L7KWB8 A0A2W1B9E8 A0A2A4KA42 A0A194QKY0 A0A2H1WK46 A0A0N1I9K7 A0A212F9C9 A0A2A4KB29 A0A194QJ23 A0A0L7KVZ8 H9JMX8 A0A023PNK9 A0A0N1IHN6 A0A194QJI7 A0A2A4JNG3 A0A0L7KJ46 A0A212F161 A0A194QPY2 A0A2A4JTG8 A0A0L7LCK3 A0A212FJ74 A0A0N0PEP7 A0A2H1W2F0 A0A0N0PEE0 A0A0N1IPS2 A0A2H1V5D8 A0A194QHU0 A0A212ENJ2 A0A2H1WVP2 A0A0L7LFR7 A0A336M7L2 A0A0N1IG19 A0A2A4JUU3 A0A2A4KAY2 A0A2H1WX63 A0A2W1BF54 A0A194QJ80 H9J017 H9J4S1 A0A2W1BG84 A0A2W1BN20 A0A336MJD0 A0A0L7KUI7 A0A0L7LHM6 A0A194QJ30 A0A2W1BD78 H9J4X7 A0A1L8DFA5 A0A0L7KLX5 A0A0N0PAH4 H9J539 A0A2H1W936 H9IV67 A0A0L7KWI6 A0A1B0CWH0 A0A1L8DF47 A0A2A4IVM9 A0A3G1T1H2 H9J4T7 A0A2H1WFL6 A0A2A4JT91 A0A2A4JKJ9 H9J4U8 A0A336MDU0 A0A139WGJ0
PDB
4LDS
E-value=4.32375e-09,
Score=147
Ontologies
GO
Topology
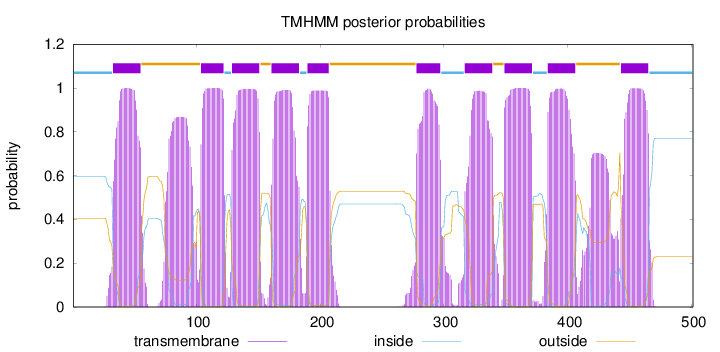
Length:
501
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
249.52486
Exp number, first 60 AAs:
22.44714
Total prob of N-in:
0.59574
POSSIBLE N-term signal
sequence
inside
1 - 32
TMhelix
33 - 55
outside
56 - 103
TMhelix
104 - 122
inside
123 - 128
TMhelix
129 - 151
outside
152 - 160
TMhelix
161 - 183
inside
184 - 189
TMhelix
190 - 207
outside
208 - 277
TMhelix
278 - 297
inside
298 - 316
TMhelix
317 - 339
outside
340 - 348
TMhelix
349 - 371
inside
372 - 383
TMhelix
384 - 406
outside
407 - 442
TMhelix
443 - 465
inside
466 - 501
Population Genetic Test Statistics
Pi
212.449257
Theta
202.107681
Tajima's D
0.973677
CLR
0.349185
CSRT
0.644417779111044
Interpretation
Uncertain
