Gene
KWMTBOMO03455
Pre Gene Modal
BGIBMGA006519
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_peroxidase-like_[Bombyx_mori]
Full name
Peroxidase
Alternative Name
Chorion peroxidase
Location in the cell
Cytoplasmic Reliability : 2.467
Sequence
CDS
ATGCAAGATTTTATGTCAACGGAGAATGTAAAAATCCTTCGAGGTCTGTACGAGTCCTTTGAGGACGTTGATCTGGTTGTTGGTGGGTCTTTGGAGCGGAATGTACCTGGAGCACAGGCTGGTCCGACATTCCTCTGCATCCTGACCGAGCAATTTTATAGAACAAGGGTCGGAGATAGATACTTCTTCGAAAACGGTGCCGACCGTGATACTGCTTTTACTCCAAGTCAATTAGAAACTATACGAAGCGGTGCATCGATGGCACGCTTGCTCTGCGACAACGCTGACGGAATCAACTTAATGCAACCGAGGGCTTTCCAGCAGATATCGCCCGGGAATCAAGTAGTATCCTGCGATGACTTACCCGCCGTGGACCTTAGCTTATGGCAGGATGCAAGTGGATTATTTAAATAA
Protein
MQDFMSTENVKILRGLYESFEDVDLVVGGSLERNVPGAQAGPTFLCILTEQFYRTRVGDRYFFENGADRDTAFTPSQLETIRSGASMARLLCDNADGINLMQPRAFQQISPGNQVVSCDDLPAVDLSLWQDASGLFK
Summary
Description
Involved in the chorion hardening process, through protein cross-linking mediated by the formation of di- and tri-tyrosine bonds.
Catalytic Activity
2 a phenolic donor + H2O2 = 2 a phenolic radical donor + 2 H2O
Cofactor
heme b
Similarity
Belongs to the peroxidase family. XPO subfamily.
Keywords
Complete proteome
Disulfide bond
Glycoprotein
Heme
Hydrogen peroxide
Iron
Metal-binding
Oxidoreductase
Peroxidase
Reference proteome
Secreted
Signal
Feature
chain Peroxidase
Uniprot
A0A2H1WWT6
A0A2A4JMJ9
A0A1Z1G6U4
A0A346D9R6
A0A088MG66
A0A194RTJ6
+ More
A0A194PXJ3 A0A194PVX4 A0A194PVR2 A0A194RPW2 A0A194RNM9 A0A194RPK5 A0A194PWT3 G9F9L1 A0A182T872 I4DR53 A0A336MK45 A0A1A9W584 B4M5J8 A0A336MHC5 A0A1B0AWK4 A0A182G7N2 A0A1A9V505 A0A1A9Z7D9 A0A1J1J076 A0A1I8NQ77 A0A1A9Y5Z1 A0A1I8NQ80 W5JNS3 B4JHE0 A0A182KV49 Q7QDZ3 A0A182WXU8 A0A182TM70 A0A182I5W9 A0A1Y9H2I7 Q16E76 A0A182PRL2 O96645 A0A1B0FFQ7 A0A182KBY5 Q177U0 A0A0L0CRA6 A0A1B6CUT2 B4NJ86 A0A0Q9WVU0 E1JIN0 Q01603 A0A0B4KHM9 A0A182VL61 A0A182FTE8 A0A0M4F792 B4PM93 B4KC82 A0A182IZD9 B4IBN7 B4QWD4 B3P3J1 A0A182Q218 A0A310SND6 B3M2B4 A0A182Y6T5 A0A1Y9IWB3 A0A182MPT8 A0A1Y9HDW6 A0A1W4VTT3 B0WM28 B0XE84 D6WCM3 A0A1B6GD63 A0A2A3EU99 A0A3B0JLT7 B5DWL8 A0A3B0JQG5 A0A088AE09 A0A0K8VYG6 A0A0L7LVA3 T1PI65 B4G4K4 A0A1W4WMG7 A0A232F2X4 K7IYZ3 A0A084V9X6 W8C2M9 A0A0A1WQI6 A0A0P4VM15 T1I7W3 A0A182KV53 A0A2J7RAM0 I1T1H0 A0A034WLG7 A0A034WQE0 A3EXQ0 A0A182P1C3 A0A182JJH9 A0A0N0BI50 A0A084V9X5 A0A182KV58 A0A0K8VYA7
A0A194PXJ3 A0A194PVX4 A0A194PVR2 A0A194RPW2 A0A194RNM9 A0A194RPK5 A0A194PWT3 G9F9L1 A0A182T872 I4DR53 A0A336MK45 A0A1A9W584 B4M5J8 A0A336MHC5 A0A1B0AWK4 A0A182G7N2 A0A1A9V505 A0A1A9Z7D9 A0A1J1J076 A0A1I8NQ77 A0A1A9Y5Z1 A0A1I8NQ80 W5JNS3 B4JHE0 A0A182KV49 Q7QDZ3 A0A182WXU8 A0A182TM70 A0A182I5W9 A0A1Y9H2I7 Q16E76 A0A182PRL2 O96645 A0A1B0FFQ7 A0A182KBY5 Q177U0 A0A0L0CRA6 A0A1B6CUT2 B4NJ86 A0A0Q9WVU0 E1JIN0 Q01603 A0A0B4KHM9 A0A182VL61 A0A182FTE8 A0A0M4F792 B4PM93 B4KC82 A0A182IZD9 B4IBN7 B4QWD4 B3P3J1 A0A182Q218 A0A310SND6 B3M2B4 A0A182Y6T5 A0A1Y9IWB3 A0A182MPT8 A0A1Y9HDW6 A0A1W4VTT3 B0WM28 B0XE84 D6WCM3 A0A1B6GD63 A0A2A3EU99 A0A3B0JLT7 B5DWL8 A0A3B0JQG5 A0A088AE09 A0A0K8VYG6 A0A0L7LVA3 T1PI65 B4G4K4 A0A1W4WMG7 A0A232F2X4 K7IYZ3 A0A084V9X6 W8C2M9 A0A0A1WQI6 A0A0P4VM15 T1I7W3 A0A182KV53 A0A2J7RAM0 I1T1H0 A0A034WLG7 A0A034WQE0 A3EXQ0 A0A182P1C3 A0A182JJH9 A0A0N0BI50 A0A084V9X5 A0A182KV58 A0A0K8VYA7
EC Number
1.11.1.7
Pubmed
25191834
26354079
22651552
17994087
26483478
20920257
+ More
23761445 20966253 12364791 17510324 11222958 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 1482687 15979004 14709175 17550304 25244985 18362917 19820115 15632085 26227816 25315136 28648823 20075255 24438588 24495485 25830018 27129103 25348373
23761445 20966253 12364791 17510324 11222958 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 1482687 15979004 14709175 17550304 25244985 18362917 19820115 15632085 26227816 25315136 28648823 20075255 24438588 24495485 25830018 27129103 25348373
EMBL
ODYU01011258
SOQ56884.1
NWSH01001024
PCG73049.1
KX244949
ARV79862.1
+ More
MH606240 AXM43807.1 KJ995807 AIN39490.1 KQ459896 KPJ19441.1 KQ459589 KPI97479.1 KPI97477.1 KPI97476.1 KPJ19444.1 KPJ19443.1 KPJ19442.1 KPI97478.1 JN033756 AEW46909.1 AK404947 BAM20393.1 UFQT01001180 SSX29359.1 CH940652 EDW58924.2 SSX29360.1 JXJN01004810 JXUM01046680 KQ561486 KXJ78429.1 CVRI01000064 CRL05204.1 ADMH02000540 ETN66032.1 CH916369 EDV93847.1 AAAB01008848 EAA07042.4 APCN01003535 CH478998 EAT32533.1 AF098717 AAC97504.1 CCAG010018629 CH477371 EAT42428.1 JRES01000032 KNC34772.1 GEDC01020121 JAS17177.1 CH964272 EDW83879.1 KRF99908.1 AE014297 ACZ94929.1 X68131 AY541497 BK002598 AGB96034.1 CP012526 ALC47695.1 CM000160 EDW96959.1 CH933806 EDW13691.2 CH480827 EDW44795.1 CM000364 EDX12625.1 CH954181 EDV48771.1 AXCN02001762 KQ760645 OAD59676.1 CH902617 EDV42305.1 AXCM01004244 DS231994 EDS30856.1 DS232811 EDS45917.1 KQ971317 EEZ99172.1 GECZ01009387 JAS60382.1 KZ288187 PBC34719.1 OUUW01000007 SPP83195.1 CM000070 EDY68158.2 SPP83193.1 GDHF01008669 JAI43645.1 JTDY01000019 KOB79392.1 KA648374 AFP63003.1 CH479179 EDW25175.1 NNAY01001189 OXU24803.1 ATLV01000829 ATLV01000830 KE523910 KFB34770.1 GAMC01010556 JAB95999.1 GBXI01013522 JAD00770.1 GDKW01003332 JAI53263.1 ACPB03011632 NEVH01006567 PNF37876.1 JN003585 AEJ88361.1 GAKP01002541 JAC56411.1 GAKP01002543 GAKP01002542 JAC56410.1 EF091968 ABN11960.1 KQ435733 KOX77231.1 KFB34769.1 GDHF01011408 GDHF01008719 JAI40906.1 JAI43595.1
MH606240 AXM43807.1 KJ995807 AIN39490.1 KQ459896 KPJ19441.1 KQ459589 KPI97479.1 KPI97477.1 KPI97476.1 KPJ19444.1 KPJ19443.1 KPJ19442.1 KPI97478.1 JN033756 AEW46909.1 AK404947 BAM20393.1 UFQT01001180 SSX29359.1 CH940652 EDW58924.2 SSX29360.1 JXJN01004810 JXUM01046680 KQ561486 KXJ78429.1 CVRI01000064 CRL05204.1 ADMH02000540 ETN66032.1 CH916369 EDV93847.1 AAAB01008848 EAA07042.4 APCN01003535 CH478998 EAT32533.1 AF098717 AAC97504.1 CCAG010018629 CH477371 EAT42428.1 JRES01000032 KNC34772.1 GEDC01020121 JAS17177.1 CH964272 EDW83879.1 KRF99908.1 AE014297 ACZ94929.1 X68131 AY541497 BK002598 AGB96034.1 CP012526 ALC47695.1 CM000160 EDW96959.1 CH933806 EDW13691.2 CH480827 EDW44795.1 CM000364 EDX12625.1 CH954181 EDV48771.1 AXCN02001762 KQ760645 OAD59676.1 CH902617 EDV42305.1 AXCM01004244 DS231994 EDS30856.1 DS232811 EDS45917.1 KQ971317 EEZ99172.1 GECZ01009387 JAS60382.1 KZ288187 PBC34719.1 OUUW01000007 SPP83195.1 CM000070 EDY68158.2 SPP83193.1 GDHF01008669 JAI43645.1 JTDY01000019 KOB79392.1 KA648374 AFP63003.1 CH479179 EDW25175.1 NNAY01001189 OXU24803.1 ATLV01000829 ATLV01000830 KE523910 KFB34770.1 GAMC01010556 JAB95999.1 GBXI01013522 JAD00770.1 GDKW01003332 JAI53263.1 ACPB03011632 NEVH01006567 PNF37876.1 JN003585 AEJ88361.1 GAKP01002541 JAC56411.1 GAKP01002543 GAKP01002542 JAC56410.1 EF091968 ABN11960.1 KQ435733 KOX77231.1 KFB34769.1 GDHF01011408 GDHF01008719 JAI40906.1 JAI43595.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000075901
UP000091820
UP000008792
+ More
UP000092460 UP000069940 UP000249989 UP000078200 UP000092445 UP000183832 UP000095300 UP000092443 UP000000673 UP000001070 UP000075882 UP000007062 UP000076407 UP000075902 UP000075840 UP000075884 UP000008820 UP000075885 UP000092444 UP000075881 UP000037069 UP000007798 UP000000803 UP000075903 UP000069272 UP000092553 UP000002282 UP000009192 UP000075880 UP000001292 UP000000304 UP000008711 UP000075886 UP000007801 UP000076408 UP000075920 UP000075883 UP000075900 UP000192221 UP000002320 UP000007266 UP000242457 UP000268350 UP000001819 UP000005203 UP000037510 UP000095301 UP000008744 UP000192223 UP000215335 UP000002358 UP000030765 UP000015103 UP000235965 UP000053105
UP000092460 UP000069940 UP000249989 UP000078200 UP000092445 UP000183832 UP000095300 UP000092443 UP000000673 UP000001070 UP000075882 UP000007062 UP000076407 UP000075902 UP000075840 UP000075884 UP000008820 UP000075885 UP000092444 UP000075881 UP000037069 UP000007798 UP000000803 UP000075903 UP000069272 UP000092553 UP000002282 UP000009192 UP000075880 UP000001292 UP000000304 UP000008711 UP000075886 UP000007801 UP000076408 UP000075920 UP000075883 UP000075900 UP000192221 UP000002320 UP000007266 UP000242457 UP000268350 UP000001819 UP000005203 UP000037510 UP000095301 UP000008744 UP000192223 UP000215335 UP000002358 UP000030765 UP000015103 UP000235965 UP000053105
PRIDE
Pfam
Interpro
IPR037120
Haem_peroxidase_sf_animal
+ More
IPR019791 Haem_peroxidase_animal
IPR029585 Pxd-like
IPR010255 Haem_peroxidase_sf
IPR013126 Hsp_70_fam
IPR011641 Tyr-kin_ephrin_A/B_rcpt-like
IPR008979 Galactose-bd-like_sf
IPR001426 Tyr_kinase_rcpt_V_CS
IPR003961 FN3_dom
IPR027936 Eph_TM
IPR009030 Growth_fac_rcpt_cys_sf
IPR013783 Ig-like_fold
IPR013761 SAM/pointed_sf
IPR001090 Ephrin_rcpt_lig-bd_dom
IPR036116 FN3_sf
IPR001660 SAM
IPR019791 Haem_peroxidase_animal
IPR029585 Pxd-like
IPR010255 Haem_peroxidase_sf
IPR013126 Hsp_70_fam
IPR011641 Tyr-kin_ephrin_A/B_rcpt-like
IPR008979 Galactose-bd-like_sf
IPR001426 Tyr_kinase_rcpt_V_CS
IPR003961 FN3_dom
IPR027936 Eph_TM
IPR009030 Growth_fac_rcpt_cys_sf
IPR013783 Ig-like_fold
IPR013761 SAM/pointed_sf
IPR001090 Ephrin_rcpt_lig-bd_dom
IPR036116 FN3_sf
IPR001660 SAM
SUPFAM
Gene 3D
CDD
ProteinModelPortal
A0A2H1WWT6
A0A2A4JMJ9
A0A1Z1G6U4
A0A346D9R6
A0A088MG66
A0A194RTJ6
+ More
A0A194PXJ3 A0A194PVX4 A0A194PVR2 A0A194RPW2 A0A194RNM9 A0A194RPK5 A0A194PWT3 G9F9L1 A0A182T872 I4DR53 A0A336MK45 A0A1A9W584 B4M5J8 A0A336MHC5 A0A1B0AWK4 A0A182G7N2 A0A1A9V505 A0A1A9Z7D9 A0A1J1J076 A0A1I8NQ77 A0A1A9Y5Z1 A0A1I8NQ80 W5JNS3 B4JHE0 A0A182KV49 Q7QDZ3 A0A182WXU8 A0A182TM70 A0A182I5W9 A0A1Y9H2I7 Q16E76 A0A182PRL2 O96645 A0A1B0FFQ7 A0A182KBY5 Q177U0 A0A0L0CRA6 A0A1B6CUT2 B4NJ86 A0A0Q9WVU0 E1JIN0 Q01603 A0A0B4KHM9 A0A182VL61 A0A182FTE8 A0A0M4F792 B4PM93 B4KC82 A0A182IZD9 B4IBN7 B4QWD4 B3P3J1 A0A182Q218 A0A310SND6 B3M2B4 A0A182Y6T5 A0A1Y9IWB3 A0A182MPT8 A0A1Y9HDW6 A0A1W4VTT3 B0WM28 B0XE84 D6WCM3 A0A1B6GD63 A0A2A3EU99 A0A3B0JLT7 B5DWL8 A0A3B0JQG5 A0A088AE09 A0A0K8VYG6 A0A0L7LVA3 T1PI65 B4G4K4 A0A1W4WMG7 A0A232F2X4 K7IYZ3 A0A084V9X6 W8C2M9 A0A0A1WQI6 A0A0P4VM15 T1I7W3 A0A182KV53 A0A2J7RAM0 I1T1H0 A0A034WLG7 A0A034WQE0 A3EXQ0 A0A182P1C3 A0A182JJH9 A0A0N0BI50 A0A084V9X5 A0A182KV58 A0A0K8VYA7
A0A194PXJ3 A0A194PVX4 A0A194PVR2 A0A194RPW2 A0A194RNM9 A0A194RPK5 A0A194PWT3 G9F9L1 A0A182T872 I4DR53 A0A336MK45 A0A1A9W584 B4M5J8 A0A336MHC5 A0A1B0AWK4 A0A182G7N2 A0A1A9V505 A0A1A9Z7D9 A0A1J1J076 A0A1I8NQ77 A0A1A9Y5Z1 A0A1I8NQ80 W5JNS3 B4JHE0 A0A182KV49 Q7QDZ3 A0A182WXU8 A0A182TM70 A0A182I5W9 A0A1Y9H2I7 Q16E76 A0A182PRL2 O96645 A0A1B0FFQ7 A0A182KBY5 Q177U0 A0A0L0CRA6 A0A1B6CUT2 B4NJ86 A0A0Q9WVU0 E1JIN0 Q01603 A0A0B4KHM9 A0A182VL61 A0A182FTE8 A0A0M4F792 B4PM93 B4KC82 A0A182IZD9 B4IBN7 B4QWD4 B3P3J1 A0A182Q218 A0A310SND6 B3M2B4 A0A182Y6T5 A0A1Y9IWB3 A0A182MPT8 A0A1Y9HDW6 A0A1W4VTT3 B0WM28 B0XE84 D6WCM3 A0A1B6GD63 A0A2A3EU99 A0A3B0JLT7 B5DWL8 A0A3B0JQG5 A0A088AE09 A0A0K8VYG6 A0A0L7LVA3 T1PI65 B4G4K4 A0A1W4WMG7 A0A232F2X4 K7IYZ3 A0A084V9X6 W8C2M9 A0A0A1WQI6 A0A0P4VM15 T1I7W3 A0A182KV53 A0A2J7RAM0 I1T1H0 A0A034WLG7 A0A034WQE0 A3EXQ0 A0A182P1C3 A0A182JJH9 A0A0N0BI50 A0A084V9X5 A0A182KV58 A0A0K8VYA7
PDB
6A4Y
E-value=6.01354e-12,
Score=164
Ontologies
GO
PANTHER
Topology
Subcellular location
Secreted
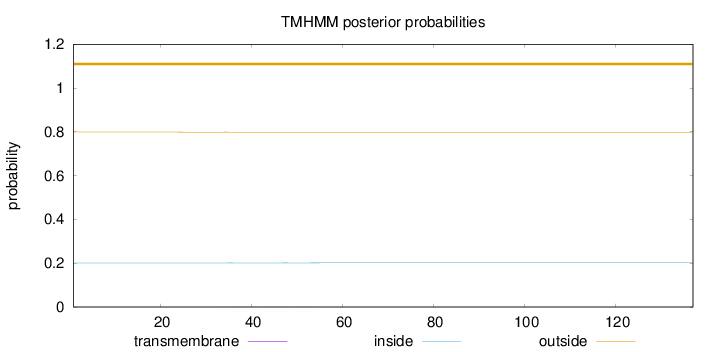
Length:
137
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01254
Exp number, first 60 AAs:
0.01254
Total prob of N-in:
0.20133
outside
1 - 137
Population Genetic Test Statistics
Pi
187.845864
Theta
30.485124
Tajima's D
2.003576
CLR
0.271689
CSRT
0.882105894705265
Interpretation
Uncertain
