Gene
KWMTBOMO03452
Pre Gene Modal
BGIBMGA006362
Annotation
PREDICTED:_mediator_of_RNA_polymerase_II_transcription_subunit_4_[Papilio_machaon]
Full name
Mediator of RNA polymerase II transcription subunit 4
Alternative Name
Mediator complex subunit 4
Location in the cell
Nuclear Reliability : 3.422
Sequence
CDS
ATGGCGTCACATTTAAGTACTAAAGATCGCTTATTGGCACTAATAGATGATATCGAACTTATTGCTAAGGAAATAATAGAAGTTTCCATAGCACCAAAACCACAGAAAGTGTCCGCTGCAGATCATGCTCAGTTGACGGACCTACTTCTATCAAAACATGGAGAATTGAAAAAAACATTAGACCTAGCTGACGAACAAGCAAAGATCCAACAGAAAATGAATGAACTCAAAGCTGAAGTAGATAGTAAGGATCAAGATATATATCACCTTCAAAGACAATTGAAAGATGCAGAACAAATTCTGGCTACATCACTGTATCAAGCTCGACAGAAACTTGCATCAATAGTGAAGTCTAGAAAGAGACCTGTACCCTCTGAGGAGTTGATCAAATTTGCACACAGAATAAGTGCATCTAATGCTGTCAGCGCACCTTTGTCGTGGCAACCTGGTGATCCTCGTCGGCCTTATCCAACTGATTTAGAAATGAGATTAGGAATGCTCGGTCGCCTCTGTGATCTGCCATTGAATGGTCACACGCCTTCTACACTGAATGAACTTCATCGTATAACAACTGGTGGAGTACCAGCTTCAGCTTCAAGCCAATTCACTTGGCATCCATCAGGCGAGCTACACATGTCTGTTGGAGGCGGTAACTCTGTAGCTATTGATGGCCGAGGGAAGGATGCTCCCAACCAAGAAGATGTTGAAGTTATGTCCACAGACTCCAGTTCAAGTTCAAGTAGTGATTCACAATGA
Protein
MASHLSTKDRLLALIDDIELIAKEIIEVSIAPKPQKVSAADHAQLTDLLLSKHGELKKTLDLADEQAKIQQKMNELKAEVDSKDQDIYHLQRQLKDAEQILATSLYQARQKLASIVKSRKRPVPSEELIKFAHRISASNAVSAPLSWQPGDPRRPYPTDLEMRLGMLGRLCDLPLNGHTPSTLNELHRITTGGVPASASSQFTWHPSGELHMSVGGGNSVAIDGRGKDAPNQEDVEVMSTDSSSSSSSDSQ
Summary
Description
Component of the Mediator complex, a coactivator involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Mediator is recruited to promoters by direct interactions with regulatory proteins and serves as a scaffold for the assembly of a functional preinitiation complex with RNA polymerase II and the general transcription factors.
Component of the Mediator complex, a coactivator involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Mediator is recruited to promoters by direct interactions with regulatory proteins and serves as a scaffold for the assembly of a functional preinitiation complex with RNA polymerase II and the general transcription factors (By similarity).
Component of the Mediator complex, a coactivator involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Mediator is recruited to promoters by direct interactions with regulatory proteins and serves as a scaffold for the assembly of a functional preinitiation complex with RNA polymerase II and the general transcription factors (By similarity).
Subunit
Component of the Mediator complex.
Similarity
Belongs to the Mediator complex subunit 4 family.
Keywords
Activator
Coiled coil
Complete proteome
Nucleus
Reference proteome
Transcription
Transcription regulation
Feature
chain Mediator of RNA polymerase II transcription subunit 4
Uniprot
H9JA17
A0A2A4JPG8
A0A194RPV8
S4NXY1
A0A2H1VEK9
A0A2Z5U7J5
+ More
A0A212F948 A0A194PVX9 A0A1B0DM83 A0A1L8DGQ3 A0A182UIA6 A0A182XUK1 A0A182VD86 A0A084VES6 A0A182JVR9 A0A182SH62 A0A182J2R8 A0A182PNI9 A0A182M6M3 A0A182L7P8 A0A182I838 Q7QH62 A0A182YF32 A0A026WY17 A0A1Y1N1D5 A0A1L8DGL1 E2B3N6 E2AIG7 A0A1Q3EYG1 E9J3B9 E0VEA4 A0A151HZ25 A0A158NIE5 A0A2J7Q6J7 A0A023EMZ6 D6WBA5 F4WJI6 A0A067QFU4 A0A2M4ARX8 W5JGL5 A0A195D316 A0A2M4BWS5 A0A0L7RDW2 A0A0N1ITD8 A0A310SNS5 B0X235 Q177A7 A0A1Q3EYF5 A0A151XDS7 V5GJM1 A0A0A9Z0Z2 A0A151IUW1 A0A0T6ATC3 A0A154PB41 A0A0P4VYZ9 A0A088AN46 R4G4N5 A0A151K185 A0A2A3EGB3 A0A023FB78 A0A2M3ZAY5 A0A182QWV9 A0A224XRV5 A0A232FFT9 K7J751 A0A182R3R1 A0A0K8VL91 A0A182NG50 W8BFP2 A0A182VX28 A0A182F8G9 A0A1B6DGF2 U4UJW5 A0A1A9XU77 A0A1A9VIB1 A0A1A9ZM46 A0A1A9WHN6 N6U1A5 A0A336MKC0 U4UKF6 U5ETT4 N6U3Y4 A0A1B6LG16 A0A1B0CG95 A0A1B6F7P7 A0A1B6I1Q0 A0A0L0BNX5 A0A1I8M6Z8 A0A1I8P5P3 A0A1J1I6I4 A0A0J7JUE7 T1IG40 A0A2S2NDS7 C4WTS2 J9K6G7 A0A2H8TNC8 A0A182GRP4 K7J0B0
A0A212F948 A0A194PVX9 A0A1B0DM83 A0A1L8DGQ3 A0A182UIA6 A0A182XUK1 A0A182VD86 A0A084VES6 A0A182JVR9 A0A182SH62 A0A182J2R8 A0A182PNI9 A0A182M6M3 A0A182L7P8 A0A182I838 Q7QH62 A0A182YF32 A0A026WY17 A0A1Y1N1D5 A0A1L8DGL1 E2B3N6 E2AIG7 A0A1Q3EYG1 E9J3B9 E0VEA4 A0A151HZ25 A0A158NIE5 A0A2J7Q6J7 A0A023EMZ6 D6WBA5 F4WJI6 A0A067QFU4 A0A2M4ARX8 W5JGL5 A0A195D316 A0A2M4BWS5 A0A0L7RDW2 A0A0N1ITD8 A0A310SNS5 B0X235 Q177A7 A0A1Q3EYF5 A0A151XDS7 V5GJM1 A0A0A9Z0Z2 A0A151IUW1 A0A0T6ATC3 A0A154PB41 A0A0P4VYZ9 A0A088AN46 R4G4N5 A0A151K185 A0A2A3EGB3 A0A023FB78 A0A2M3ZAY5 A0A182QWV9 A0A224XRV5 A0A232FFT9 K7J751 A0A182R3R1 A0A0K8VL91 A0A182NG50 W8BFP2 A0A182VX28 A0A182F8G9 A0A1B6DGF2 U4UJW5 A0A1A9XU77 A0A1A9VIB1 A0A1A9ZM46 A0A1A9WHN6 N6U1A5 A0A336MKC0 U4UKF6 U5ETT4 N6U3Y4 A0A1B6LG16 A0A1B0CG95 A0A1B6F7P7 A0A1B6I1Q0 A0A0L0BNX5 A0A1I8M6Z8 A0A1I8P5P3 A0A1J1I6I4 A0A0J7JUE7 T1IG40 A0A2S2NDS7 C4WTS2 J9K6G7 A0A2H8TNC8 A0A182GRP4 K7J0B0
Pubmed
19121390
26354079
23622113
22118469
24438588
20966253
+ More
12364791 25244985 24508170 30249741 28004739 20798317 21282665 20566863 21347285 24945155 18362917 19820115 21719571 24845553 20920257 23761445 17510324 25401762 26823975 27129103 25474469 28648823 20075255 24495485 23537049 26108605 25315136 26483478
12364791 25244985 24508170 30249741 28004739 20798317 21282665 20566863 21347285 24945155 18362917 19820115 21719571 24845553 20920257 23761445 17510324 25401762 26823975 27129103 25474469 28648823 20075255 24495485 23537049 26108605 25315136 26483478
EMBL
BABH01009332
NWSH01000938
PCG73474.1
KQ459896
KPJ19439.1
GAIX01011977
+ More
JAA80583.1 ODYU01001883 SOQ38714.1 AP017538 BBB06936.1 AGBW02009653 OWR50229.1 KQ459589 KPI97482.1 AJVK01006897 GFDF01008448 JAV05636.1 ATLV01012290 KE524778 KFB36470.1 AXCM01002109 APCN01002617 AAAB01008817 KK107063 QOIP01000005 EZA60970.1 RLU22546.1 GEZM01020153 GEZM01020152 JAV89357.1 GFDF01008486 JAV05598.1 GL445365 EFN89692.1 GL439791 EFN66757.1 GFDL01014719 JAV20326.1 GL768061 EFZ12691.1 AAZO01001504 DS235088 EEB11710.1 KQ976704 KYM77083.1 ADTU01016738 NEVH01017470 PNF24198.1 GAPW01003353 JAC10245.1 KQ971312 EEZ98716.2 GL888183 EGI65642.1 KK853582 KDR06509.1 GGFK01010234 MBW43555.1 ADMH02001471 ETN62463.1 KQ976973 KYN06754.1 GGFJ01008368 MBW57509.1 KQ414613 KOC68921.1 KQ435824 KOX72224.1 KQ761768 OAD57128.1 DS232279 EDS38965.1 CH477379 GFDL01014710 JAV20335.1 KQ982294 KYQ58408.1 GALX01006649 JAB61817.1 GBHO01008119 GBRD01013700 GDHC01016036 JAG35485.1 JAG52126.1 JAQ02593.1 KQ980946 KYN11235.1 LJIG01022876 KRT78278.1 KQ434864 KZC09027.1 GDKW01001816 JAI54779.1 GAHY01000621 JAA76889.1 KQ981178 KYN45281.1 KZ288269 PBC30041.1 GBBI01000534 JAC18178.1 GGFM01004922 MBW25673.1 AXCN02001190 GFTR01003868 JAW12558.1 NNAY01000297 OXU29380.1 AAZX01008222 GDHF01013004 JAI39310.1 GAMC01014464 GAMC01014462 GAMC01014460 JAB92091.1 GEDC01012535 JAS24763.1 KB632237 ERL90481.1 APGK01043586 KB741015 ENN75330.1 UFQT01001195 SSX29429.1 ERL90480.1 GANO01002641 JAB57230.1 ENN75331.1 GEBQ01017377 JAT22600.1 AJWK01010865 AJWK01010866 GECZ01023491 JAS46278.1 GECU01026870 JAS80836.1 JRES01001582 KNC21757.1 CVRI01000039 CRK94550.1 LBMM01030588 KMQ81888.1 ACPB03002685 GGMR01002493 MBY15112.1 AK340745 BAH71292.1 ABLF02040145 ABLF02040154 GFXV01003868 MBW15673.1 JXUM01016808 KQ560466 KXJ82280.1
JAA80583.1 ODYU01001883 SOQ38714.1 AP017538 BBB06936.1 AGBW02009653 OWR50229.1 KQ459589 KPI97482.1 AJVK01006897 GFDF01008448 JAV05636.1 ATLV01012290 KE524778 KFB36470.1 AXCM01002109 APCN01002617 AAAB01008817 KK107063 QOIP01000005 EZA60970.1 RLU22546.1 GEZM01020153 GEZM01020152 JAV89357.1 GFDF01008486 JAV05598.1 GL445365 EFN89692.1 GL439791 EFN66757.1 GFDL01014719 JAV20326.1 GL768061 EFZ12691.1 AAZO01001504 DS235088 EEB11710.1 KQ976704 KYM77083.1 ADTU01016738 NEVH01017470 PNF24198.1 GAPW01003353 JAC10245.1 KQ971312 EEZ98716.2 GL888183 EGI65642.1 KK853582 KDR06509.1 GGFK01010234 MBW43555.1 ADMH02001471 ETN62463.1 KQ976973 KYN06754.1 GGFJ01008368 MBW57509.1 KQ414613 KOC68921.1 KQ435824 KOX72224.1 KQ761768 OAD57128.1 DS232279 EDS38965.1 CH477379 GFDL01014710 JAV20335.1 KQ982294 KYQ58408.1 GALX01006649 JAB61817.1 GBHO01008119 GBRD01013700 GDHC01016036 JAG35485.1 JAG52126.1 JAQ02593.1 KQ980946 KYN11235.1 LJIG01022876 KRT78278.1 KQ434864 KZC09027.1 GDKW01001816 JAI54779.1 GAHY01000621 JAA76889.1 KQ981178 KYN45281.1 KZ288269 PBC30041.1 GBBI01000534 JAC18178.1 GGFM01004922 MBW25673.1 AXCN02001190 GFTR01003868 JAW12558.1 NNAY01000297 OXU29380.1 AAZX01008222 GDHF01013004 JAI39310.1 GAMC01014464 GAMC01014462 GAMC01014460 JAB92091.1 GEDC01012535 JAS24763.1 KB632237 ERL90481.1 APGK01043586 KB741015 ENN75330.1 UFQT01001195 SSX29429.1 ERL90480.1 GANO01002641 JAB57230.1 ENN75331.1 GEBQ01017377 JAT22600.1 AJWK01010865 AJWK01010866 GECZ01023491 JAS46278.1 GECU01026870 JAS80836.1 JRES01001582 KNC21757.1 CVRI01000039 CRK94550.1 LBMM01030588 KMQ81888.1 ACPB03002685 GGMR01002493 MBY15112.1 AK340745 BAH71292.1 ABLF02040145 ABLF02040154 GFXV01003868 MBW15673.1 JXUM01016808 KQ560466 KXJ82280.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000092462
+ More
UP000075902 UP000076407 UP000075903 UP000030765 UP000075881 UP000075901 UP000075880 UP000075885 UP000075883 UP000075882 UP000075840 UP000007062 UP000076408 UP000053097 UP000279307 UP000008237 UP000000311 UP000009046 UP000078540 UP000005205 UP000235965 UP000007266 UP000007755 UP000027135 UP000000673 UP000078542 UP000053825 UP000053105 UP000002320 UP000008820 UP000075809 UP000078492 UP000076502 UP000005203 UP000078541 UP000242457 UP000075886 UP000215335 UP000002358 UP000075900 UP000075884 UP000075920 UP000069272 UP000030742 UP000092443 UP000078200 UP000092445 UP000091820 UP000019118 UP000092461 UP000037069 UP000095301 UP000095300 UP000183832 UP000036403 UP000015103 UP000007819 UP000069940 UP000249989
UP000075902 UP000076407 UP000075903 UP000030765 UP000075881 UP000075901 UP000075880 UP000075885 UP000075883 UP000075882 UP000075840 UP000007062 UP000076408 UP000053097 UP000279307 UP000008237 UP000000311 UP000009046 UP000078540 UP000005205 UP000235965 UP000007266 UP000007755 UP000027135 UP000000673 UP000078542 UP000053825 UP000053105 UP000002320 UP000008820 UP000075809 UP000078492 UP000076502 UP000005203 UP000078541 UP000242457 UP000075886 UP000215335 UP000002358 UP000075900 UP000075884 UP000075920 UP000069272 UP000030742 UP000092443 UP000078200 UP000092445 UP000091820 UP000019118 UP000092461 UP000037069 UP000095301 UP000095300 UP000183832 UP000036403 UP000015103 UP000007819 UP000069940 UP000249989
PRIDE
Pfam
Interpro
IPR019258
Mediator_Med4
+ More
IPR038914 DCAF15
IPR032734 DCAF15_WD40
IPR001680 WD40_repeat
IPR007109 Brix
IPR017986 WD40_repeat_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR013979 TIF_beta_prop-like
IPR019775 WD40_repeat_CS
IPR020472 G-protein_beta_WD-40_rep
IPR011425 Med9
IPR005146 B3/B4_tRNA-bd
IPR004531 Phe-tRNA-synth_IIc_bsu_arc
IPR040659 PhetRS_B1
IPR005147 tRNA_synthase_B5-dom
IPR020825 Phe-tRNA_synthase_B3/B4
IPR009061 DNA-bd_dom_put_sf
IPR041616 PheRS_beta_core
IPR038914 DCAF15
IPR032734 DCAF15_WD40
IPR001680 WD40_repeat
IPR007109 Brix
IPR017986 WD40_repeat_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR013979 TIF_beta_prop-like
IPR019775 WD40_repeat_CS
IPR020472 G-protein_beta_WD-40_rep
IPR011425 Med9
IPR005146 B3/B4_tRNA-bd
IPR004531 Phe-tRNA-synth_IIc_bsu_arc
IPR040659 PhetRS_B1
IPR005147 tRNA_synthase_B5-dom
IPR020825 Phe-tRNA_synthase_B3/B4
IPR009061 DNA-bd_dom_put_sf
IPR041616 PheRS_beta_core
Gene 3D
ProteinModelPortal
H9JA17
A0A2A4JPG8
A0A194RPV8
S4NXY1
A0A2H1VEK9
A0A2Z5U7J5
+ More
A0A212F948 A0A194PVX9 A0A1B0DM83 A0A1L8DGQ3 A0A182UIA6 A0A182XUK1 A0A182VD86 A0A084VES6 A0A182JVR9 A0A182SH62 A0A182J2R8 A0A182PNI9 A0A182M6M3 A0A182L7P8 A0A182I838 Q7QH62 A0A182YF32 A0A026WY17 A0A1Y1N1D5 A0A1L8DGL1 E2B3N6 E2AIG7 A0A1Q3EYG1 E9J3B9 E0VEA4 A0A151HZ25 A0A158NIE5 A0A2J7Q6J7 A0A023EMZ6 D6WBA5 F4WJI6 A0A067QFU4 A0A2M4ARX8 W5JGL5 A0A195D316 A0A2M4BWS5 A0A0L7RDW2 A0A0N1ITD8 A0A310SNS5 B0X235 Q177A7 A0A1Q3EYF5 A0A151XDS7 V5GJM1 A0A0A9Z0Z2 A0A151IUW1 A0A0T6ATC3 A0A154PB41 A0A0P4VYZ9 A0A088AN46 R4G4N5 A0A151K185 A0A2A3EGB3 A0A023FB78 A0A2M3ZAY5 A0A182QWV9 A0A224XRV5 A0A232FFT9 K7J751 A0A182R3R1 A0A0K8VL91 A0A182NG50 W8BFP2 A0A182VX28 A0A182F8G9 A0A1B6DGF2 U4UJW5 A0A1A9XU77 A0A1A9VIB1 A0A1A9ZM46 A0A1A9WHN6 N6U1A5 A0A336MKC0 U4UKF6 U5ETT4 N6U3Y4 A0A1B6LG16 A0A1B0CG95 A0A1B6F7P7 A0A1B6I1Q0 A0A0L0BNX5 A0A1I8M6Z8 A0A1I8P5P3 A0A1J1I6I4 A0A0J7JUE7 T1IG40 A0A2S2NDS7 C4WTS2 J9K6G7 A0A2H8TNC8 A0A182GRP4 K7J0B0
A0A212F948 A0A194PVX9 A0A1B0DM83 A0A1L8DGQ3 A0A182UIA6 A0A182XUK1 A0A182VD86 A0A084VES6 A0A182JVR9 A0A182SH62 A0A182J2R8 A0A182PNI9 A0A182M6M3 A0A182L7P8 A0A182I838 Q7QH62 A0A182YF32 A0A026WY17 A0A1Y1N1D5 A0A1L8DGL1 E2B3N6 E2AIG7 A0A1Q3EYG1 E9J3B9 E0VEA4 A0A151HZ25 A0A158NIE5 A0A2J7Q6J7 A0A023EMZ6 D6WBA5 F4WJI6 A0A067QFU4 A0A2M4ARX8 W5JGL5 A0A195D316 A0A2M4BWS5 A0A0L7RDW2 A0A0N1ITD8 A0A310SNS5 B0X235 Q177A7 A0A1Q3EYF5 A0A151XDS7 V5GJM1 A0A0A9Z0Z2 A0A151IUW1 A0A0T6ATC3 A0A154PB41 A0A0P4VYZ9 A0A088AN46 R4G4N5 A0A151K185 A0A2A3EGB3 A0A023FB78 A0A2M3ZAY5 A0A182QWV9 A0A224XRV5 A0A232FFT9 K7J751 A0A182R3R1 A0A0K8VL91 A0A182NG50 W8BFP2 A0A182VX28 A0A182F8G9 A0A1B6DGF2 U4UJW5 A0A1A9XU77 A0A1A9VIB1 A0A1A9ZM46 A0A1A9WHN6 N6U1A5 A0A336MKC0 U4UKF6 U5ETT4 N6U3Y4 A0A1B6LG16 A0A1B0CG95 A0A1B6F7P7 A0A1B6I1Q0 A0A0L0BNX5 A0A1I8M6Z8 A0A1I8P5P3 A0A1J1I6I4 A0A0J7JUE7 T1IG40 A0A2S2NDS7 C4WTS2 J9K6G7 A0A2H8TNC8 A0A182GRP4 K7J0B0
Ontologies
GO
Topology
Subcellular location
Nucleus
Length:
251
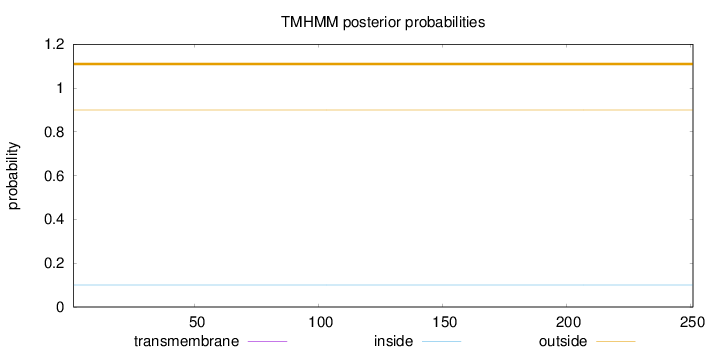
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00017
Exp number, first 60 AAs:
0
Total prob of N-in:
0.10083
outside
1 - 251
Population Genetic Test Statistics
Pi
226.396934
Theta
212.225197
Tajima's D
0.485445
CLR
0.034161
CSRT
0.509424528773561
Interpretation
Uncertain
