Gene
KWMTBOMO03449
Pre Gene Modal
BGIBMGA006516
Annotation
PREDICTED:_protein_lap4_isoform_X3_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.291
Sequence
CDS
ATGTTTCGATGTATACCTTTGTTCAAATGTAATCGCCAAGTGGAAAGTGTAGACAAGCGGCACTGTTCGTTGCCAACAGTACCTGAAGACATTTTGCGGTATTCTAGGAGCTTGGAAGAACTATTACTGGATGCTAATCATATAAGAGATTTACCTAAGAATTTCTTCAGATTACATCGATTAAGGCGACTTGGTCTCAGTGATAATGAGATACACAAACTGCCAGCTGATATCCAGAATTTTGAAAATTTAGTAGAATTGGATGTATCACGCAATGACATTCCGGATATACCTGAGGACATCAAGAAGCTGCGTGCCCTCCAGATCGCGGACTTCAGCAGCAATCCTATACCAAGGCTGCCGGCCGGCTTCAGTCAACTTCGGGCCCTTACGGTCCTCGGCCTCAATGATATGTCTCTTACTAGTTTACCCAACGATTTCGGCAGTTTAGTGTCACTTCAATCTTTGGAATTGCGAGAAAACCTACTTAAGTCACTGCCAGAGTCCTTGAAGAATTTGACCAATCTCCAGAGACTAGATTTAGGTGATAATGAAATTGAGGAATTGCCAGGGTTTATTGGAGAGCTTCCTGCTTTAGAAGAACTTTGGTTGGACCATAACAAGTTGCAGTATTTGCCGCCTGAGATCGGCAATCTCAAAGCGCTGATCTGCTTAGATGTTAGTGAGAACAAATTAGAGAAGATACCAGAAGAAATTGGCGGACTTTCATCATTAACCGACTTGCACCTCTCCCAAAACATGTTAGAATCCATACCTGATGGTATAGGTCATTTATACAAGTTAACTATACTGAAGTTGGACCAAAATAGATTACATACATTAAACGAAAACGTTGGAAGGTGTACAAATCTACAAGAGCTTATATTAACGGAGAATTTTTTAACGGAATTGCCTGACTCTATTGGAAATCTCAACAAGTTGACCGTTTTGAATGTAGATAGAAATTCGTTAGCCGAAATTCCGATGGATATCGGTAATATGACGCTGTTAGGGGTCCTGAGTTTAAGAGATAATAAATTGACGAAACTGCCGAACGAATTAGGGAACTGTACGTCACTTCACGTTTTGGACGTAAGCGGAAATCGGTTGCAATACCTTCCGTACACGCTGGTAAATTTGGAACTAAAAGCGGTGTGGTTGTCCGAAAACCAAGCGCAACCACTCTTGACTTTCCAAACGGACAGAGACGAAAGCACCGGGGAGAACGTCCTTACTTGTTTCCTCTTACCGCAGCTCAGTTACACTCAACAACCAGATACGAGCGTTTCCCGCGATCGTGATTCGGACAACGAAGATTGGGAGGAGAAGGAGGCGTCGCGCACTCATTCCGTTAAGTTCACGGAAGACGTTTCGGACGTTCGAGAGACACCGTTCGTAAGACAGAACACGCCTCATCCGAAAGACTTGAAAGCGAAAGCTCACAAGCTTTTGGCTGGACGATCACCGGACAATGCGATCACGCAACCAGCGAAGACCGAATCGCCCACTACGAACGGATTACCTTTGTCCCCTATAAAAATTGTTCCAGTCGTAGAAGAGAAAGTAGAGGTGAGTTTATTTAAAACAAGCGGCCCGCTCCGGCTCCGCTCGGGTCTTTAA
Protein
MFRCIPLFKCNRQVESVDKRHCSLPTVPEDILRYSRSLEELLLDANHIRDLPKNFFRLHRLRRLGLSDNEIHKLPADIQNFENLVELDVSRNDIPDIPEDIKKLRALQIADFSSNPIPRLPAGFSQLRALTVLGLNDMSLTSLPNDFGSLVSLQSLELRENLLKSLPESLKNLTNLQRLDLGDNEIEELPGFIGELPALEELWLDHNKLQYLPPEIGNLKALICLDVSENKLEKIPEEIGGLSSLTDLHLSQNMLESIPDGIGHLYKLTILKLDQNRLHTLNENVGRCTNLQELILTENFLTELPDSIGNLNKLTVLNVDRNSLAEIPMDIGNMTLLGVLSLRDNKLTKLPNELGNCTSLHVLDVSGNRLQYLPYTLVNLELKAVWLSENQAQPLLTFQTDRDESTGENVLTCFLLPQLSYTQQPDTSVSRDRDSDNEDWEEKEASRTHSVKFTEDVSDVRETPFVRQNTPHPKDLKAKAHKLLAGRSPDNAITQPAKTESPTTNGLPLSPIKIVPVVEEKVEVSLFKTSGPLRLRSGL
Summary
Uniprot
A0A2A4JNV8
A0A2A4JPU3
A0A2A4JN06
A0A2A4JN43
A0A2A4JNU0
A0A2A4JN16
+ More
A0A2A4JPY7 A0A2A4JP37 A0A2A4JPX6 A0A2A4JN33 A0A2A4JNC7 A0A194RTJ1 A0A194Q253 A0A1L8DIR0 A0A1Q3EUR3 A0A069DXR8 A0A151WRI2 A0A151IG73 A0A195BAM7 A0A0V0G5C8 A0A195FKD1 F4WRA6 E2BMS1 A0A0K8TGI4 A0A0K8TGU1 A0A0K8TAR0 A0A0K8TGI0 A0A0A9XBF3 A0A0A9XAE9 A0A0A9XCS6 A0A0A9XK01 A0A0K8TG82 A0A0K8TBV1 A0A0A9XBE8 A0A0K8TAS1 A0A0A9Z136 A0A0K8THH0 A0A0A9XIP3 A0A0K8TB62 A0A0K8TG76 A0A0K8TAU0 A0A0A9X8C7 A0A158NUU3 E2AD26 A0A336M751 A0A146L5J4 A0A146L9V0 A0A336LYR4 A0A336KKD6 V9IC56 A0A154PAZ8 A0A088ANG2 A0A1B6KH23 A0A1B6KKP4 A0A1B6LUV6 A0A1B6KAM2 A0A1B6H6Z5 A0A034VDB5 A0A0A1XP83 W8C7R8 A0A0K8VK50 A0A1B6LTJ2 A0A1B6MQT7 A0A1B6LBT3 A0A1B6MHB9 A0A0K8U4C6 V5GMN3 A0A1Y1LNA5 A0A1B6IV05 A0A1Y1LS62 A0A026WS96 A0A1Y1LNA2 A0A1Y1LN58 A0A2M3YXI9 A0A3L8DFK9 A0A0K8V903 A0A1Y1LNA7 A0A0Q9X030 A0A2M3YXK4 A0A2M4CKD5 A0A1Y1LN88 A0A0K8UD34 T1PJ26 A0A0M3QXL6 K7J1A0 A0A1J1HWG7 B4JGP0 A0A2M3YYE9 A0A336LXZ6 A0A336LUY7 A0A336KF30 A0A1J1HWT6 A0A0P8Y0R0 A0A0P8Y0S7 A0A1B6DBB0 E1JIZ0 E1JIZ1 A0A0P9A0Z3 A0A1W4V7Z8 E1JIZ2
A0A2A4JPY7 A0A2A4JP37 A0A2A4JPX6 A0A2A4JN33 A0A2A4JNC7 A0A194RTJ1 A0A194Q253 A0A1L8DIR0 A0A1Q3EUR3 A0A069DXR8 A0A151WRI2 A0A151IG73 A0A195BAM7 A0A0V0G5C8 A0A195FKD1 F4WRA6 E2BMS1 A0A0K8TGI4 A0A0K8TGU1 A0A0K8TAR0 A0A0K8TGI0 A0A0A9XBF3 A0A0A9XAE9 A0A0A9XCS6 A0A0A9XK01 A0A0K8TG82 A0A0K8TBV1 A0A0A9XBE8 A0A0K8TAS1 A0A0A9Z136 A0A0K8THH0 A0A0A9XIP3 A0A0K8TB62 A0A0K8TG76 A0A0K8TAU0 A0A0A9X8C7 A0A158NUU3 E2AD26 A0A336M751 A0A146L5J4 A0A146L9V0 A0A336LYR4 A0A336KKD6 V9IC56 A0A154PAZ8 A0A088ANG2 A0A1B6KH23 A0A1B6KKP4 A0A1B6LUV6 A0A1B6KAM2 A0A1B6H6Z5 A0A034VDB5 A0A0A1XP83 W8C7R8 A0A0K8VK50 A0A1B6LTJ2 A0A1B6MQT7 A0A1B6LBT3 A0A1B6MHB9 A0A0K8U4C6 V5GMN3 A0A1Y1LNA5 A0A1B6IV05 A0A1Y1LS62 A0A026WS96 A0A1Y1LNA2 A0A1Y1LN58 A0A2M3YXI9 A0A3L8DFK9 A0A0K8V903 A0A1Y1LNA7 A0A0Q9X030 A0A2M3YXK4 A0A2M4CKD5 A0A1Y1LN88 A0A0K8UD34 T1PJ26 A0A0M3QXL6 K7J1A0 A0A1J1HWG7 B4JGP0 A0A2M3YYE9 A0A336LXZ6 A0A336LUY7 A0A336KF30 A0A1J1HWT6 A0A0P8Y0R0 A0A0P8Y0S7 A0A1B6DBB0 E1JIZ0 E1JIZ1 A0A0P9A0Z3 A0A1W4V7Z8 E1JIZ2
Pubmed
EMBL
NWSH01000938
PCG73456.1
PCG73463.1
PCG73455.1
PCG73467.1
PCG73469.1
+ More
PCG73465.1 PCG73472.1 PCG73468.1 PCG73460.1 PCG73457.1 PCG73471.1 KQ459896 KPJ19436.1 KQ459589 KPI97485.1 GFDF01007752 JAV06332.1 GFDL01016013 JAV19032.1 GBGD01000066 JAC88823.1 KQ982806 KYQ50460.1 KQ977739 KYN00224.1 KQ976532 KYM81593.1 GECL01002867 JAP03257.1 KQ981512 KYN40816.1 GL888284 EGI63323.1 GL449319 EFN83009.1 GBRD01001222 JAG64599.1 GBRD01001225 JAG64596.1 GBRD01003179 JAG62642.1 GBRD01001227 JAG64594.1 GBHO01026623 JAG16981.1 GBHO01026625 JAG16979.1 GBHO01026158 JAG17446.1 GBHO01026159 JAG17445.1 GBRD01001223 JAG64598.1 GBRD01003181 JAG62640.1 GBHO01026628 JAG16976.1 GBRD01003178 JAG62643.1 GBHO01008079 JAG35525.1 GBRD01001226 JAG64595.1 GBHO01026629 JAG16975.1 GBRD01003180 JAG62641.1 GBRD01001224 JAG64597.1 GBRD01003182 JAG62639.1 GBHO01026627 JAG16977.1 ADTU01026648 ADTU01026649 ADTU01026650 GL438658 EFN68662.1 UFQS01000217 UFQT01000217 SSX01478.1 SSX21858.1 GDHC01015724 JAQ02905.1 GDHC01014264 JAQ04365.1 SSX01479.1 SSX21859.1 SSX01480.1 SSX21860.1 JR038395 AEY58237.1 KQ434864 KZC08987.1 GEBQ01029209 JAT10768.1 GEBQ01027949 JAT12028.1 GEBQ01012515 JAT27462.1 GEBQ01031474 JAT08503.1 GECU01037313 JAS70393.1 GAKP01018448 JAC40504.1 GBXI01001934 JAD12358.1 GAMC01007866 JAB98689.1 GDHF01013065 JAI39249.1 GEBQ01013038 JAT26939.1 GEBQ01001708 JAT38269.1 GEBQ01018841 JAT21136.1 GEBQ01004664 JAT35313.1 GDHF01030755 JAI21559.1 GALX01003107 JAB65359.1 GEZM01051169 JAV75103.1 GECU01016948 JAS90758.1 GEZM01051163 JAV75110.1 KK107119 EZA58536.1 GEZM01051167 GEZM01051162 JAV75112.1 GEZM01051168 JAV75104.1 GGFM01000228 MBW20979.1 QOIP01000009 RLU18689.1 GDHF01016957 JAI35357.1 GEZM01051164 JAV75109.1 CH933806 KRG01484.1 GGFM01000205 MBW20956.1 GGFL01001541 MBW65719.1 GEZM01051170 JAV75102.1 GDHF01027836 JAI24478.1 KA648684 AFP63313.1 CP012526 ALC46143.1 CVRI01000028 CRK92447.1 CH916369 EDV92644.1 GGFM01000548 MBW21299.1 SSX01481.1 SSX21861.1 SSX01482.1 SSX21862.1 SSX01483.1 SSX21863.1 CRK92451.1 CH902617 KPU80360.1 KPU80378.1 GEDC01014319 JAS22979.1 AE014297 ACZ95040.1 ACZ95039.1 KPU80374.1 ACZ95041.1
PCG73465.1 PCG73472.1 PCG73468.1 PCG73460.1 PCG73457.1 PCG73471.1 KQ459896 KPJ19436.1 KQ459589 KPI97485.1 GFDF01007752 JAV06332.1 GFDL01016013 JAV19032.1 GBGD01000066 JAC88823.1 KQ982806 KYQ50460.1 KQ977739 KYN00224.1 KQ976532 KYM81593.1 GECL01002867 JAP03257.1 KQ981512 KYN40816.1 GL888284 EGI63323.1 GL449319 EFN83009.1 GBRD01001222 JAG64599.1 GBRD01001225 JAG64596.1 GBRD01003179 JAG62642.1 GBRD01001227 JAG64594.1 GBHO01026623 JAG16981.1 GBHO01026625 JAG16979.1 GBHO01026158 JAG17446.1 GBHO01026159 JAG17445.1 GBRD01001223 JAG64598.1 GBRD01003181 JAG62640.1 GBHO01026628 JAG16976.1 GBRD01003178 JAG62643.1 GBHO01008079 JAG35525.1 GBRD01001226 JAG64595.1 GBHO01026629 JAG16975.1 GBRD01003180 JAG62641.1 GBRD01001224 JAG64597.1 GBRD01003182 JAG62639.1 GBHO01026627 JAG16977.1 ADTU01026648 ADTU01026649 ADTU01026650 GL438658 EFN68662.1 UFQS01000217 UFQT01000217 SSX01478.1 SSX21858.1 GDHC01015724 JAQ02905.1 GDHC01014264 JAQ04365.1 SSX01479.1 SSX21859.1 SSX01480.1 SSX21860.1 JR038395 AEY58237.1 KQ434864 KZC08987.1 GEBQ01029209 JAT10768.1 GEBQ01027949 JAT12028.1 GEBQ01012515 JAT27462.1 GEBQ01031474 JAT08503.1 GECU01037313 JAS70393.1 GAKP01018448 JAC40504.1 GBXI01001934 JAD12358.1 GAMC01007866 JAB98689.1 GDHF01013065 JAI39249.1 GEBQ01013038 JAT26939.1 GEBQ01001708 JAT38269.1 GEBQ01018841 JAT21136.1 GEBQ01004664 JAT35313.1 GDHF01030755 JAI21559.1 GALX01003107 JAB65359.1 GEZM01051169 JAV75103.1 GECU01016948 JAS90758.1 GEZM01051163 JAV75110.1 KK107119 EZA58536.1 GEZM01051167 GEZM01051162 JAV75112.1 GEZM01051168 JAV75104.1 GGFM01000228 MBW20979.1 QOIP01000009 RLU18689.1 GDHF01016957 JAI35357.1 GEZM01051164 JAV75109.1 CH933806 KRG01484.1 GGFM01000205 MBW20956.1 GGFL01001541 MBW65719.1 GEZM01051170 JAV75102.1 GDHF01027836 JAI24478.1 KA648684 AFP63313.1 CP012526 ALC46143.1 CVRI01000028 CRK92447.1 CH916369 EDV92644.1 GGFM01000548 MBW21299.1 SSX01481.1 SSX21861.1 SSX01482.1 SSX21862.1 SSX01483.1 SSX21863.1 CRK92451.1 CH902617 KPU80360.1 KPU80378.1 GEDC01014319 JAS22979.1 AE014297 ACZ95040.1 ACZ95039.1 KPU80374.1 ACZ95041.1
Proteomes
Interpro
SUPFAM
SSF50156
SSF50156
Gene 3D
ProteinModelPortal
A0A2A4JNV8
A0A2A4JPU3
A0A2A4JN06
A0A2A4JN43
A0A2A4JNU0
A0A2A4JN16
+ More
A0A2A4JPY7 A0A2A4JP37 A0A2A4JPX6 A0A2A4JN33 A0A2A4JNC7 A0A194RTJ1 A0A194Q253 A0A1L8DIR0 A0A1Q3EUR3 A0A069DXR8 A0A151WRI2 A0A151IG73 A0A195BAM7 A0A0V0G5C8 A0A195FKD1 F4WRA6 E2BMS1 A0A0K8TGI4 A0A0K8TGU1 A0A0K8TAR0 A0A0K8TGI0 A0A0A9XBF3 A0A0A9XAE9 A0A0A9XCS6 A0A0A9XK01 A0A0K8TG82 A0A0K8TBV1 A0A0A9XBE8 A0A0K8TAS1 A0A0A9Z136 A0A0K8THH0 A0A0A9XIP3 A0A0K8TB62 A0A0K8TG76 A0A0K8TAU0 A0A0A9X8C7 A0A158NUU3 E2AD26 A0A336M751 A0A146L5J4 A0A146L9V0 A0A336LYR4 A0A336KKD6 V9IC56 A0A154PAZ8 A0A088ANG2 A0A1B6KH23 A0A1B6KKP4 A0A1B6LUV6 A0A1B6KAM2 A0A1B6H6Z5 A0A034VDB5 A0A0A1XP83 W8C7R8 A0A0K8VK50 A0A1B6LTJ2 A0A1B6MQT7 A0A1B6LBT3 A0A1B6MHB9 A0A0K8U4C6 V5GMN3 A0A1Y1LNA5 A0A1B6IV05 A0A1Y1LS62 A0A026WS96 A0A1Y1LNA2 A0A1Y1LN58 A0A2M3YXI9 A0A3L8DFK9 A0A0K8V903 A0A1Y1LNA7 A0A0Q9X030 A0A2M3YXK4 A0A2M4CKD5 A0A1Y1LN88 A0A0K8UD34 T1PJ26 A0A0M3QXL6 K7J1A0 A0A1J1HWG7 B4JGP0 A0A2M3YYE9 A0A336LXZ6 A0A336LUY7 A0A336KF30 A0A1J1HWT6 A0A0P8Y0R0 A0A0P8Y0S7 A0A1B6DBB0 E1JIZ0 E1JIZ1 A0A0P9A0Z3 A0A1W4V7Z8 E1JIZ2
A0A2A4JPY7 A0A2A4JP37 A0A2A4JPX6 A0A2A4JN33 A0A2A4JNC7 A0A194RTJ1 A0A194Q253 A0A1L8DIR0 A0A1Q3EUR3 A0A069DXR8 A0A151WRI2 A0A151IG73 A0A195BAM7 A0A0V0G5C8 A0A195FKD1 F4WRA6 E2BMS1 A0A0K8TGI4 A0A0K8TGU1 A0A0K8TAR0 A0A0K8TGI0 A0A0A9XBF3 A0A0A9XAE9 A0A0A9XCS6 A0A0A9XK01 A0A0K8TG82 A0A0K8TBV1 A0A0A9XBE8 A0A0K8TAS1 A0A0A9Z136 A0A0K8THH0 A0A0A9XIP3 A0A0K8TB62 A0A0K8TG76 A0A0K8TAU0 A0A0A9X8C7 A0A158NUU3 E2AD26 A0A336M751 A0A146L5J4 A0A146L9V0 A0A336LYR4 A0A336KKD6 V9IC56 A0A154PAZ8 A0A088ANG2 A0A1B6KH23 A0A1B6KKP4 A0A1B6LUV6 A0A1B6KAM2 A0A1B6H6Z5 A0A034VDB5 A0A0A1XP83 W8C7R8 A0A0K8VK50 A0A1B6LTJ2 A0A1B6MQT7 A0A1B6LBT3 A0A1B6MHB9 A0A0K8U4C6 V5GMN3 A0A1Y1LNA5 A0A1B6IV05 A0A1Y1LS62 A0A026WS96 A0A1Y1LNA2 A0A1Y1LN58 A0A2M3YXI9 A0A3L8DFK9 A0A0K8V903 A0A1Y1LNA7 A0A0Q9X030 A0A2M3YXK4 A0A2M4CKD5 A0A1Y1LN88 A0A0K8UD34 T1PJ26 A0A0M3QXL6 K7J1A0 A0A1J1HWG7 B4JGP0 A0A2M3YYE9 A0A336LXZ6 A0A336LUY7 A0A336KF30 A0A1J1HWT6 A0A0P8Y0R0 A0A0P8Y0S7 A0A1B6DBB0 E1JIZ0 E1JIZ1 A0A0P9A0Z3 A0A1W4V7Z8 E1JIZ2
PDB
4U08
E-value=2.94347e-41,
Score=425
Ontologies
GO
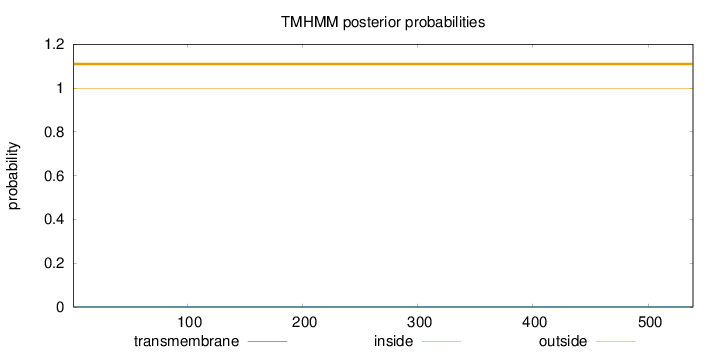
Topology
Length:
539
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00084
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00069
outside
1 - 539
Population Genetic Test Statistics
Pi
210.645766
Theta
188.409209
Tajima's D
0.445698
CLR
0.111059
CSRT
0.504074796260187
Interpretation
Uncertain
