Gene
KWMTBOMO03448
Pre Gene Modal
BGIBMGA006364
Annotation
PREDICTED:_pyrroline-5-carboxylate_reductase_[Bombyx_mori]
Full name
Pyrroline-5-carboxylate reductase
Location in the cell
PlasmaMembrane Reliability : 0.863
Sequence
CDS
ATGAGTAATAATTTTAAGATCGGTTTTGTTGGAGGAGGGAATATGTCAACAGCAATTGTTCAAGGAATTTTAAAAAATGATGCTTATCCGGCTTCAAATATTTGGGTTTCTGGGCCACACTTGGAAAACCTACAGAAATGGTCACTTTTAAACACAAATGTTAGCATAAAAAACTCTGATGTTGTTAACAACTGTGAAATAATATTTCTAGGCATAAAGCCAAATAAATTAGAAACAGCTGTGCAACAATGTCTAAATGGCATAGAATCTCTTAGTAAGAAAAAATCAATATTGTTTGTTTCTATGTTGGCTGGAAGGAAAATAAATGATTTGCAGGAGGTGTTAAGTGTACTATCTCCTTATGCTGATATAAGAATAATAAGGATAATGCCAAATACACCTATGGCCGTAGGGGCTGGTATATGTCTTTACACAACGGATAACAATGTTTCACAAGATCAATGTAAATTTTTGGAAGATCTATTGAGTAGCACTGCACTATGTGAAAGAGTTGAAGAACCAACAATGGACACACTTGGAGTTTTGACTGCTTGTGGACCTGCTTTTATTTATATTGTGATAGAAGCCTTAGCCGATGGTGCTGTGAAGCAAGGTGTTCCTCGAGCAATGGCATTGCGTCACGCAGCACAGGTGGTAGTGGGTAGCGGTCAGATGGTCTTGCAAACCGGTAAACATCCTGGTCTATTGAAAGATGAAGTTTGTTCTCCTGCTGGTTCCACTATATGTGGTGTGACTGAGTTAGAAAATGGAAGATTGAGGTCTACATTCATAAATGCAATTGAAGCTGCAACAAATAGGACGAAAGATCTTGGAAAACGTTAA
Protein
MSNNFKIGFVGGGNMSTAIVQGILKNDAYPASNIWVSGPHLENLQKWSLLNTNVSIKNSDVVNNCEIIFLGIKPNKLETAVQQCLNGIESLSKKKSILFVSMLAGRKINDLQEVLSVLSPYADIRIIRIMPNTPMAVGAGICLYTTDNNVSQDQCKFLEDLLSSTALCERVEEPTMDTLGVLTACGPAFIYIVIEALADGAVKQGVPRAMALRHAAQVVVGSGQMVLQTGKHPGLLKDEVCSPAGSTICGVTELENGRLRSTFINAIEAATNRTKDLGKR
Summary
Catalytic Activity
L-proline + NADP(+) = 1-pyrroline-5-carboxylate + 2 H(+) + NADPH
Similarity
Belongs to the pyrroline-5-carboxylate reductase family.
Uniprot
A0A2H1W8R1
A0A194PVS3
A0A2A4JPD9
A0A194RQ37
A0A0N0PF62
V5SJ26
+ More
T1GKW2 A0A0K8U594 A0A0K8WHL9 B4LXA3 A0A1W4UBH3 B4JSG3 A0A034VUN4 W8BLL8 B4K7A8 A0A2A3E4E4 A0A0B4J2M9 T1PI37 B4PMQ9 A0A154PDA8 A0A232EF11 A0A1I8MXR4 A0A1L8EDU4 Q9V3F8 A0A1L8EBN6 K7IZW8 O96643 A0A0L7QPP0 B4ILV5 Q6NP47 A0A3B0K3N1 A0A1I8PIP8 A0A0T6AU53 B3P380 Q29BT4 B4GPN9 A0A0J7KT06 A0A1A9XI54 A0A195CSA1 V5GFX0 B4NHY8 A0A195FSU1 F4WFL5 A0A3L8DWK8 A0A151WIA5 A0A151J4M5 A0A1A9VR32 D3TNW1 A0A151HZ37 Q0QHK4 D6W9S1 E2AAJ1 B3LVB6 A0A158NSH6 A0A0M4EN63 A0A1B0BWJ5 A0A1B0D902 A0A1B0ACL7 A0A1A9X186 W8CBR5 A0A2J7QRD1 A0A1L8E2P1 A0A1Z5L5M3 A0A336K9D3 A0A0K8SXU3 A0A1W4WUK0 A0A146KRB4 A0A182QY18 A0A026WNB1 E0VE13 A0A0K8SX01 A0A0P4VNQ5 A0A0K8TLB1 A0A1Y1M0D2 A0A182NG99 A0A023F1E3 A0A182GF55 A0A224XU55 N6TZQ5 A0A023ENV0 A0A293MXX1 A0A182IYG7 A0A0T6ATZ6 B0WCX9 Q2KQA2 A0A1Q3FCU0 A0A182JZB4 Q1HRH9 A0A0A9Z490 Q16YV4 A0A2M4BVP5 A0A2R5LJX3 A0A084VCD2 A0A0V0GAN1 W5JGW5 A0A182R762 A0A1B6IKB3 A0A1B6MFT9 A0A1J1I4U9
T1GKW2 A0A0K8U594 A0A0K8WHL9 B4LXA3 A0A1W4UBH3 B4JSG3 A0A034VUN4 W8BLL8 B4K7A8 A0A2A3E4E4 A0A0B4J2M9 T1PI37 B4PMQ9 A0A154PDA8 A0A232EF11 A0A1I8MXR4 A0A1L8EDU4 Q9V3F8 A0A1L8EBN6 K7IZW8 O96643 A0A0L7QPP0 B4ILV5 Q6NP47 A0A3B0K3N1 A0A1I8PIP8 A0A0T6AU53 B3P380 Q29BT4 B4GPN9 A0A0J7KT06 A0A1A9XI54 A0A195CSA1 V5GFX0 B4NHY8 A0A195FSU1 F4WFL5 A0A3L8DWK8 A0A151WIA5 A0A151J4M5 A0A1A9VR32 D3TNW1 A0A151HZ37 Q0QHK4 D6W9S1 E2AAJ1 B3LVB6 A0A158NSH6 A0A0M4EN63 A0A1B0BWJ5 A0A1B0D902 A0A1B0ACL7 A0A1A9X186 W8CBR5 A0A2J7QRD1 A0A1L8E2P1 A0A1Z5L5M3 A0A336K9D3 A0A0K8SXU3 A0A1W4WUK0 A0A146KRB4 A0A182QY18 A0A026WNB1 E0VE13 A0A0K8SX01 A0A0P4VNQ5 A0A0K8TLB1 A0A1Y1M0D2 A0A182NG99 A0A023F1E3 A0A182GF55 A0A224XU55 N6TZQ5 A0A023ENV0 A0A293MXX1 A0A182IYG7 A0A0T6ATZ6 B0WCX9 Q2KQA2 A0A1Q3FCU0 A0A182JZB4 Q1HRH9 A0A0A9Z490 Q16YV4 A0A2M4BVP5 A0A2R5LJX3 A0A084VCD2 A0A0V0GAN1 W5JGW5 A0A182R762 A0A1B6IKB3 A0A1B6MFT9 A0A1J1I4U9
EC Number
1.5.1.2
Pubmed
26354079
17994087
25348373
24495485
17550304
28648823
+ More
25315136 10858562 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20075255 15632085 21719571 30249741 20353571 18362917 19820115 20798317 21347285 28528879 26823975 24508170 20566863 27129103 26369729 28004739 25474469 26483478 23537049 24945155 15804581 17204158 25401762 17510324 24438588 20920257 23761445
25315136 10858562 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20075255 15632085 21719571 30249741 20353571 18362917 19820115 20798317 21347285 28528879 26823975 24508170 20566863 27129103 26369729 28004739 25474469 26483478 23537049 24945155 15804581 17204158 25401762 17510324 24438588 20920257 23761445
EMBL
ODYU01006743
SOQ48874.1
KQ459589
KPI97486.1
NWSH01000938
PCG73454.1
+ More
KQ459896 KPJ19435.1 LADJ01005682 KPJ20884.1 KF647644 AHB50506.1 CAQQ02390338 GDHF01030500 JAI21814.1 GDHF01001755 JAI50559.1 CH940650 EDW66755.1 CH916373 EDV94703.1 GAKP01013157 GAKP01013155 JAC45797.1 GAMC01004605 GAMC01004603 JAC01951.1 CH933806 EDW14232.1 KZ288416 PBC26029.1 KA647800 AFP62429.1 CM000160 EDW95979.1 KQ434878 KZC09896.1 NNAY01005138 OXU16959.1 GFDG01001937 JAV16862.1 AF170829 AE014297 AAD49740.1 AAF55626.1 GFDG01002773 JAV16026.1 AF098020 AAC70780.1 KQ414815 KOC60524.1 CH480892 EDW55431.1 BT011084 AAR82750.1 OUUW01000007 SPP82550.1 LJIG01022798 KRT78657.1 CH954181 EDV48532.1 CM000070 EAL26913.1 CH479186 EDW39122.1 LBMM01003504 KMQ93456.1 KQ977329 KYN03571.1 GALX01005592 JAB62874.1 CH964272 EDW83638.1 KQ981276 KYN43511.1 GL888120 EGI66992.1 QOIP01000003 RLU24831.1 KQ983089 KYQ47568.1 KQ980159 KYN17429.1 EZ423113 ADD19389.1 KQ976719 KYM76752.1 DQ374015 ABG77993.1 KQ971312 EEZ98115.1 GL438128 EFN69538.1 CH902617 EDV43648.1 ADTU01024930 ADTU01024931 CP012526 ALC46064.1 JXJN01021835 AJVK01027986 AJVK01027987 GAMC01004606 JAC01950.1 NEVH01011914 PNF31136.1 GFDF01001168 JAV12916.1 GFJQ02004679 JAW02291.1 UFQS01000141 UFQT01000141 SSX00412.1 SSX20792.1 GBRD01008086 JAG57735.1 GDHC01020703 GDHC01008850 JAP97925.1 JAQ09779.1 AXCN02001627 KK107161 EZA56589.1 DS235088 EEB11619.1 GBRD01008087 JAG57734.1 GDKW01000286 JAI56309.1 GDAI01002652 JAI14951.1 GEZM01047934 JAV76687.1 GBBI01003604 JAC15108.1 JXUM01059245 KQ562042 KXJ76834.1 GFTR01004793 JAW11633.1 APGK01054985 KB741257 KB631615 ENN71742.1 ERL84683.1 JXUM01039862 JXUM01039863 GAPW01002972 KQ561220 JAC10626.1 KXJ79312.1 GFWV01012305 GFWV01019472 MAA44200.1 KRT78656.1 DS231891 EDS43939.1 AY623401 AAV31910.1 GFDL01009685 JAV25360.1 DQ440115 ABF18148.1 GBHO01035610 GBHO01025400 GBHO01004290 GBHO01004288 GBHO01004287 GBHO01004286 GBHO01004285 GBHO01004282 GBHO01002798 GBHO01002796 GBHO01002795 GBHO01002794 GDHC01018697 GDHC01016435 GDHC01015029 GDHC01005258 JAG07994.1 JAG18204.1 JAG39314.1 JAG39316.1 JAG39317.1 JAG39318.1 JAG39319.1 JAG39322.1 JAG40806.1 JAG40808.1 JAG40809.1 JAG40810.1 JAP99931.1 JAQ02194.1 JAQ03600.1 JAQ13371.1 CH477508 EAT39807.1 GGFJ01008004 MBW57145.1 GGLE01005714 MBY09840.1 ATLV01010660 KE524596 KFB35626.1 GECL01001661 JAP04463.1 ADMH02001428 ETN62563.1 GECU01020346 GECU01010791 JAS87360.1 JAS96915.1 GEBQ01010665 GEBQ01005201 GEBQ01003774 JAT29312.1 JAT34776.1 JAT36203.1 CVRI01000039 CRK94620.1
KQ459896 KPJ19435.1 LADJ01005682 KPJ20884.1 KF647644 AHB50506.1 CAQQ02390338 GDHF01030500 JAI21814.1 GDHF01001755 JAI50559.1 CH940650 EDW66755.1 CH916373 EDV94703.1 GAKP01013157 GAKP01013155 JAC45797.1 GAMC01004605 GAMC01004603 JAC01951.1 CH933806 EDW14232.1 KZ288416 PBC26029.1 KA647800 AFP62429.1 CM000160 EDW95979.1 KQ434878 KZC09896.1 NNAY01005138 OXU16959.1 GFDG01001937 JAV16862.1 AF170829 AE014297 AAD49740.1 AAF55626.1 GFDG01002773 JAV16026.1 AF098020 AAC70780.1 KQ414815 KOC60524.1 CH480892 EDW55431.1 BT011084 AAR82750.1 OUUW01000007 SPP82550.1 LJIG01022798 KRT78657.1 CH954181 EDV48532.1 CM000070 EAL26913.1 CH479186 EDW39122.1 LBMM01003504 KMQ93456.1 KQ977329 KYN03571.1 GALX01005592 JAB62874.1 CH964272 EDW83638.1 KQ981276 KYN43511.1 GL888120 EGI66992.1 QOIP01000003 RLU24831.1 KQ983089 KYQ47568.1 KQ980159 KYN17429.1 EZ423113 ADD19389.1 KQ976719 KYM76752.1 DQ374015 ABG77993.1 KQ971312 EEZ98115.1 GL438128 EFN69538.1 CH902617 EDV43648.1 ADTU01024930 ADTU01024931 CP012526 ALC46064.1 JXJN01021835 AJVK01027986 AJVK01027987 GAMC01004606 JAC01950.1 NEVH01011914 PNF31136.1 GFDF01001168 JAV12916.1 GFJQ02004679 JAW02291.1 UFQS01000141 UFQT01000141 SSX00412.1 SSX20792.1 GBRD01008086 JAG57735.1 GDHC01020703 GDHC01008850 JAP97925.1 JAQ09779.1 AXCN02001627 KK107161 EZA56589.1 DS235088 EEB11619.1 GBRD01008087 JAG57734.1 GDKW01000286 JAI56309.1 GDAI01002652 JAI14951.1 GEZM01047934 JAV76687.1 GBBI01003604 JAC15108.1 JXUM01059245 KQ562042 KXJ76834.1 GFTR01004793 JAW11633.1 APGK01054985 KB741257 KB631615 ENN71742.1 ERL84683.1 JXUM01039862 JXUM01039863 GAPW01002972 KQ561220 JAC10626.1 KXJ79312.1 GFWV01012305 GFWV01019472 MAA44200.1 KRT78656.1 DS231891 EDS43939.1 AY623401 AAV31910.1 GFDL01009685 JAV25360.1 DQ440115 ABF18148.1 GBHO01035610 GBHO01025400 GBHO01004290 GBHO01004288 GBHO01004287 GBHO01004286 GBHO01004285 GBHO01004282 GBHO01002798 GBHO01002796 GBHO01002795 GBHO01002794 GDHC01018697 GDHC01016435 GDHC01015029 GDHC01005258 JAG07994.1 JAG18204.1 JAG39314.1 JAG39316.1 JAG39317.1 JAG39318.1 JAG39319.1 JAG39322.1 JAG40806.1 JAG40808.1 JAG40809.1 JAG40810.1 JAP99931.1 JAQ02194.1 JAQ03600.1 JAQ13371.1 CH477508 EAT39807.1 GGFJ01008004 MBW57145.1 GGLE01005714 MBY09840.1 ATLV01010660 KE524596 KFB35626.1 GECL01001661 JAP04463.1 ADMH02001428 ETN62563.1 GECU01020346 GECU01010791 JAS87360.1 JAS96915.1 GEBQ01010665 GEBQ01005201 GEBQ01003774 JAT29312.1 JAT34776.1 JAT36203.1 CVRI01000039 CRK94620.1
Proteomes
UP000053268
UP000218220
UP000053240
UP000015102
UP000008792
UP000192221
+ More
UP000001070 UP000009192 UP000242457 UP000005203 UP000002282 UP000076502 UP000215335 UP000095301 UP000000803 UP000002358 UP000053825 UP000001292 UP000268350 UP000095300 UP000008711 UP000001819 UP000008744 UP000036403 UP000092443 UP000078542 UP000007798 UP000078541 UP000007755 UP000279307 UP000075809 UP000078492 UP000078200 UP000078540 UP000007266 UP000000311 UP000007801 UP000005205 UP000092553 UP000092460 UP000092462 UP000092445 UP000091820 UP000235965 UP000192223 UP000075886 UP000053097 UP000009046 UP000075884 UP000069940 UP000249989 UP000019118 UP000030742 UP000075880 UP000002320 UP000075881 UP000008820 UP000030765 UP000000673 UP000075900 UP000183832
UP000001070 UP000009192 UP000242457 UP000005203 UP000002282 UP000076502 UP000215335 UP000095301 UP000000803 UP000002358 UP000053825 UP000001292 UP000268350 UP000095300 UP000008711 UP000001819 UP000008744 UP000036403 UP000092443 UP000078542 UP000007798 UP000078541 UP000007755 UP000279307 UP000075809 UP000078492 UP000078200 UP000078540 UP000007266 UP000000311 UP000007801 UP000005205 UP000092553 UP000092460 UP000092462 UP000092445 UP000091820 UP000235965 UP000192223 UP000075886 UP000053097 UP000009046 UP000075884 UP000069940 UP000249989 UP000019118 UP000030742 UP000075880 UP000002320 UP000075881 UP000008820 UP000030765 UP000000673 UP000075900 UP000183832
Interpro
ProteinModelPortal
A0A2H1W8R1
A0A194PVS3
A0A2A4JPD9
A0A194RQ37
A0A0N0PF62
V5SJ26
+ More
T1GKW2 A0A0K8U594 A0A0K8WHL9 B4LXA3 A0A1W4UBH3 B4JSG3 A0A034VUN4 W8BLL8 B4K7A8 A0A2A3E4E4 A0A0B4J2M9 T1PI37 B4PMQ9 A0A154PDA8 A0A232EF11 A0A1I8MXR4 A0A1L8EDU4 Q9V3F8 A0A1L8EBN6 K7IZW8 O96643 A0A0L7QPP0 B4ILV5 Q6NP47 A0A3B0K3N1 A0A1I8PIP8 A0A0T6AU53 B3P380 Q29BT4 B4GPN9 A0A0J7KT06 A0A1A9XI54 A0A195CSA1 V5GFX0 B4NHY8 A0A195FSU1 F4WFL5 A0A3L8DWK8 A0A151WIA5 A0A151J4M5 A0A1A9VR32 D3TNW1 A0A151HZ37 Q0QHK4 D6W9S1 E2AAJ1 B3LVB6 A0A158NSH6 A0A0M4EN63 A0A1B0BWJ5 A0A1B0D902 A0A1B0ACL7 A0A1A9X186 W8CBR5 A0A2J7QRD1 A0A1L8E2P1 A0A1Z5L5M3 A0A336K9D3 A0A0K8SXU3 A0A1W4WUK0 A0A146KRB4 A0A182QY18 A0A026WNB1 E0VE13 A0A0K8SX01 A0A0P4VNQ5 A0A0K8TLB1 A0A1Y1M0D2 A0A182NG99 A0A023F1E3 A0A182GF55 A0A224XU55 N6TZQ5 A0A023ENV0 A0A293MXX1 A0A182IYG7 A0A0T6ATZ6 B0WCX9 Q2KQA2 A0A1Q3FCU0 A0A182JZB4 Q1HRH9 A0A0A9Z490 Q16YV4 A0A2M4BVP5 A0A2R5LJX3 A0A084VCD2 A0A0V0GAN1 W5JGW5 A0A182R762 A0A1B6IKB3 A0A1B6MFT9 A0A1J1I4U9
T1GKW2 A0A0K8U594 A0A0K8WHL9 B4LXA3 A0A1W4UBH3 B4JSG3 A0A034VUN4 W8BLL8 B4K7A8 A0A2A3E4E4 A0A0B4J2M9 T1PI37 B4PMQ9 A0A154PDA8 A0A232EF11 A0A1I8MXR4 A0A1L8EDU4 Q9V3F8 A0A1L8EBN6 K7IZW8 O96643 A0A0L7QPP0 B4ILV5 Q6NP47 A0A3B0K3N1 A0A1I8PIP8 A0A0T6AU53 B3P380 Q29BT4 B4GPN9 A0A0J7KT06 A0A1A9XI54 A0A195CSA1 V5GFX0 B4NHY8 A0A195FSU1 F4WFL5 A0A3L8DWK8 A0A151WIA5 A0A151J4M5 A0A1A9VR32 D3TNW1 A0A151HZ37 Q0QHK4 D6W9S1 E2AAJ1 B3LVB6 A0A158NSH6 A0A0M4EN63 A0A1B0BWJ5 A0A1B0D902 A0A1B0ACL7 A0A1A9X186 W8CBR5 A0A2J7QRD1 A0A1L8E2P1 A0A1Z5L5M3 A0A336K9D3 A0A0K8SXU3 A0A1W4WUK0 A0A146KRB4 A0A182QY18 A0A026WNB1 E0VE13 A0A0K8SX01 A0A0P4VNQ5 A0A0K8TLB1 A0A1Y1M0D2 A0A182NG99 A0A023F1E3 A0A182GF55 A0A224XU55 N6TZQ5 A0A023ENV0 A0A293MXX1 A0A182IYG7 A0A0T6ATZ6 B0WCX9 Q2KQA2 A0A1Q3FCU0 A0A182JZB4 Q1HRH9 A0A0A9Z490 Q16YV4 A0A2M4BVP5 A0A2R5LJX3 A0A084VCD2 A0A0V0GAN1 W5JGW5 A0A182R762 A0A1B6IKB3 A0A1B6MFT9 A0A1J1I4U9
PDB
2GRA
E-value=5.10767e-42,
Score=428
Ontologies
PATHWAY
GO
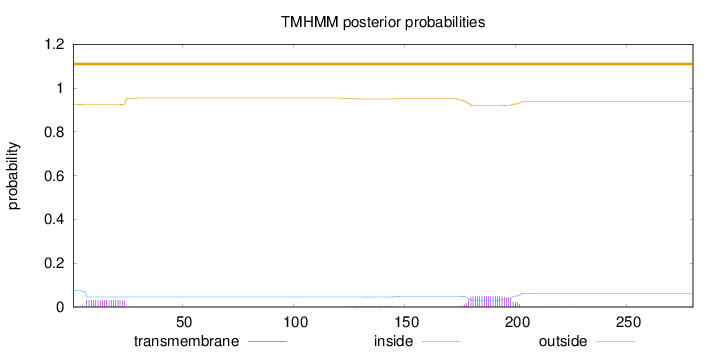
Topology
Length:
280
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.73104
Exp number, first 60 AAs:
0.572
Total prob of N-in:
0.07548
outside
1 - 280
Population Genetic Test Statistics
Pi
190.201095
Theta
172.370326
Tajima's D
0.667433
CLR
139.221713
CSRT
0.566571671416429
Interpretation
Uncertain
