Gene
KWMTBOMO03442
Annotation
PREDICTED:_proton-coupled_folate_transporter-like_[Papilio_machaon]
Full name
Angiotensin-converting enzyme
Location in the cell
PlasmaMembrane Reliability : 4.379
Sequence
CDS
ATGAACATTTCATCTCTTCTGTACGTGCAATTCGTAATGCAAGATGTGAATGTTAACACCGAAAACCGAGTGTCGGTGTTTAATTGGAAGCTACCATTGAAGGCGTTCAAAAGCCTAATAAAGAGCCGGGAAGGACACAAAAGGAAAATAATATTACTCATGTTGCTTGTGTCTCTCGGAGATAGAGTTCTACTATCAGCTGAGGTGCTAATCGCCTACATGTACTATAGGTACAAATTTCATTGGGACGACATAATGTTTGGCTCGTTTTTGGCATATAAGAACATCATCAGCTTCTTTGGTACATTATTAATATTGAGTCTGCTGAAGCGACGCTTGAAACTGTCGGACGAGATGGTAGGAGTTTTCAGCTGCCTGTCAAACATATTATCTACATCTGGTCTCATTGCCACCACTTACACTTACTGCGTATTTGTGCTTCCTTTGCTCGGGATAATATCTCAAGGATCTCAGGTCGTCCAGAGGCCGCTACTGAATAAGCAAATTTTACCAACTGAACAAGGTAAAATATACAGCGTTATGGGAGCCCTCCAATCTGCAATACAGACATTGTCGTCGCCGCTATACTCCCTGTTATACAGCAGAACTGTCTCAACGTTTCCAGATGCTTGGCTTCTGCCGGGATTGGGACTCGCGTTAATACAATTAACAGCATACCTAATAGCAAGGAGGTTACGTCTGAAATCAAATGTCGCAGACAATGATAATATGAAAGTTAATACTGAAGATTGTTGTTTAGATATAGAAGACAAAGTGATTAAAAATGATCTCAACTCTGTCGAATTAGCGTCTGAAAAGTGTGCCGAAAAGTAA
Protein
MNISSLLYVQFVMQDVNVNTENRVSVFNWKLPLKAFKSLIKSREGHKRKIILLMLLVSLGDRVLLSAEVLIAYMYYRYKFHWDDIMFGSFLAYKNIISFFGTLLILSLLKRRLKLSDEMVGVFSCLSNILSTSGLIATTYTYCVFVLPLLGIISQGSQVVQRPLLNKQILPTEQGKIYSVMGALQSAIQTLSSPLYSLLYSRTVSTFPDAWLLPGLGLALIQLTAYLIARRLRLKSNVADNDNMKVNTEDCCLDIEDKVIKNDLNSVELASEKCAEK
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M2 family.
Belongs to the adenylyl cyclase class-4/guanylyl cyclase family.
Belongs to the adenylyl cyclase class-4/guanylyl cyclase family.
Uniprot
A0A194RPU8
A0A1E1WVC4
A0A212F393
A0A194PVY9
A0A0L7KYA7
A0A1B6LLW9
+ More
A0A1B6CE26 A0A194QS69 A0A194QAI0 A0A1E1W1C8 A0A146M6W4 A0A0A9W4N6 A0A182QCE8 A0A2H1W938 A0A182J2E5 A0A2W1BZD6 A0A182N9U1 A0A084WT83 Q16W79 A0A2M4BIV5 A0A2M4AAV7 A0A2M3Z330 A0A2M4BIX1 A0A182W3L5 A0A2M3Z363 A0A2M4CTY3 A0A182MSS2 A0A2M4CUE7 A0A2M4CSN5 A0A2M4CUE3 A0A182VHT3 Q5TSW5 A0A182PQS5 A0A2C9H8S5 A0A194RE89 A0A2S2NH79 A0A182RDY9 A0A182H1C2 A0A212FAM1 A0A194PXP8 A0A182K2U1 A0A1J1HPT6 A0A1Y1KH29 A0A1Q3FLB8 A0A1Q3FLC7 A0A2S2NX57 A0A2H1WT49 B0WAI1 A0A1Q3FKN7 A0A1Q3FL61
A0A1B6CE26 A0A194QS69 A0A194QAI0 A0A1E1W1C8 A0A146M6W4 A0A0A9W4N6 A0A182QCE8 A0A2H1W938 A0A182J2E5 A0A2W1BZD6 A0A182N9U1 A0A084WT83 Q16W79 A0A2M4BIV5 A0A2M4AAV7 A0A2M3Z330 A0A2M4BIX1 A0A182W3L5 A0A2M3Z363 A0A2M4CTY3 A0A182MSS2 A0A2M4CUE7 A0A2M4CSN5 A0A2M4CUE3 A0A182VHT3 Q5TSW5 A0A182PQS5 A0A2C9H8S5 A0A194RE89 A0A2S2NH79 A0A182RDY9 A0A182H1C2 A0A212FAM1 A0A194PXP8 A0A182K2U1 A0A1J1HPT6 A0A1Y1KH29 A0A1Q3FLB8 A0A1Q3FLC7 A0A2S2NX57 A0A2H1WT49 B0WAI1 A0A1Q3FKN7 A0A1Q3FL61
EC Number
3.4.-.-
Pubmed
EMBL
KQ459896
KPJ19429.1
GDQN01000298
JAT90756.1
AGBW02010586
OWR48209.1
+ More
KQ459589 KPI97492.1 JTDY01004458 KOB68135.1 GEBQ01015246 JAT24731.1 GEDC01025663 JAS11635.1 KQ461155 KPJ08368.1 KQ459249 KPJ02439.1 GDQN01010353 JAT80701.1 GDHC01003145 JAQ15484.1 GBHO01041233 GBRD01004096 JAG02371.1 JAG61725.1 AXCN02001200 ODYU01007104 SOQ49580.1 KZ149899 PZC78617.1 ATLV01026865 ATLV01026866 KE525420 KFB53427.1 CH477573 EAT38831.1 GGFJ01003793 MBW52934.1 GGFK01004578 MBW37899.1 GGFM01002176 MBW22927.1 GGFJ01003792 MBW52933.1 GGFM01002194 MBW22945.1 GGFL01004110 MBW68288.1 AXCM01000458 GGFL01004789 MBW68967.1 GGFL01004111 MBW68289.1 GGFL01004788 MBW68966.1 AAAB01008898 EAL40562.4 KQ460323 KPJ15907.1 GGMR01003934 MBY16553.1 JXUM01102960 KQ564767 KXJ71770.1 AGBW02009461 OWR50763.1 KQ459586 KPI98097.1 CVRI01000006 CRK88225.1 GEZM01088643 GEZM01088642 JAV58047.1 GFDL01006678 JAV28367.1 GFDL01006731 JAV28314.1 GGMR01009161 MBY21780.1 ODYU01010862 SOQ56231.1 DS231872 EDS41404.1 GFDL01006875 JAV28170.1 GFDL01006801 JAV28244.1
KQ459589 KPI97492.1 JTDY01004458 KOB68135.1 GEBQ01015246 JAT24731.1 GEDC01025663 JAS11635.1 KQ461155 KPJ08368.1 KQ459249 KPJ02439.1 GDQN01010353 JAT80701.1 GDHC01003145 JAQ15484.1 GBHO01041233 GBRD01004096 JAG02371.1 JAG61725.1 AXCN02001200 ODYU01007104 SOQ49580.1 KZ149899 PZC78617.1 ATLV01026865 ATLV01026866 KE525420 KFB53427.1 CH477573 EAT38831.1 GGFJ01003793 MBW52934.1 GGFK01004578 MBW37899.1 GGFM01002176 MBW22927.1 GGFJ01003792 MBW52933.1 GGFM01002194 MBW22945.1 GGFL01004110 MBW68288.1 AXCM01000458 GGFL01004789 MBW68967.1 GGFL01004111 MBW68289.1 GGFL01004788 MBW68966.1 AAAB01008898 EAL40562.4 KQ460323 KPJ15907.1 GGMR01003934 MBY16553.1 JXUM01102960 KQ564767 KXJ71770.1 AGBW02009461 OWR50763.1 KQ459586 KPI98097.1 CVRI01000006 CRK88225.1 GEZM01088643 GEZM01088642 JAV58047.1 GFDL01006678 JAV28367.1 GFDL01006731 JAV28314.1 GGMR01009161 MBY21780.1 ODYU01010862 SOQ56231.1 DS231872 EDS41404.1 GFDL01006875 JAV28170.1 GFDL01006801 JAV28244.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A194RPU8
A0A1E1WVC4
A0A212F393
A0A194PVY9
A0A0L7KYA7
A0A1B6LLW9
+ More
A0A1B6CE26 A0A194QS69 A0A194QAI0 A0A1E1W1C8 A0A146M6W4 A0A0A9W4N6 A0A182QCE8 A0A2H1W938 A0A182J2E5 A0A2W1BZD6 A0A182N9U1 A0A084WT83 Q16W79 A0A2M4BIV5 A0A2M4AAV7 A0A2M3Z330 A0A2M4BIX1 A0A182W3L5 A0A2M3Z363 A0A2M4CTY3 A0A182MSS2 A0A2M4CUE7 A0A2M4CSN5 A0A2M4CUE3 A0A182VHT3 Q5TSW5 A0A182PQS5 A0A2C9H8S5 A0A194RE89 A0A2S2NH79 A0A182RDY9 A0A182H1C2 A0A212FAM1 A0A194PXP8 A0A182K2U1 A0A1J1HPT6 A0A1Y1KH29 A0A1Q3FLB8 A0A1Q3FLC7 A0A2S2NX57 A0A2H1WT49 B0WAI1 A0A1Q3FKN7 A0A1Q3FL61
A0A1B6CE26 A0A194QS69 A0A194QAI0 A0A1E1W1C8 A0A146M6W4 A0A0A9W4N6 A0A182QCE8 A0A2H1W938 A0A182J2E5 A0A2W1BZD6 A0A182N9U1 A0A084WT83 Q16W79 A0A2M4BIV5 A0A2M4AAV7 A0A2M3Z330 A0A2M4BIX1 A0A182W3L5 A0A2M3Z363 A0A2M4CTY3 A0A182MSS2 A0A2M4CUE7 A0A2M4CSN5 A0A2M4CUE3 A0A182VHT3 Q5TSW5 A0A182PQS5 A0A2C9H8S5 A0A194RE89 A0A2S2NH79 A0A182RDY9 A0A182H1C2 A0A212FAM1 A0A194PXP8 A0A182K2U1 A0A1J1HPT6 A0A1Y1KH29 A0A1Q3FLB8 A0A1Q3FLC7 A0A2S2NX57 A0A2H1WT49 B0WAI1 A0A1Q3FKN7 A0A1Q3FL61
Ontologies
GO
PANTHER
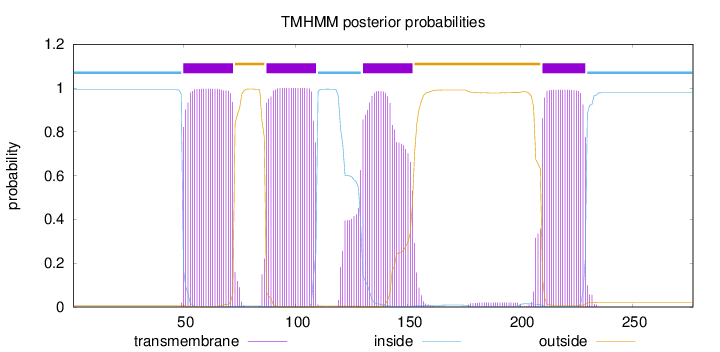
Topology
Length:
277
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
91.00208
Exp number, first 60 AAs:
10.61674
Total prob of N-in:
0.99212
POSSIBLE N-term signal
sequence
inside
1 - 49
TMhelix
50 - 72
outside
73 - 86
TMhelix
87 - 109
inside
110 - 129
TMhelix
130 - 152
outside
153 - 209
TMhelix
210 - 229
inside
230 - 277
Population Genetic Test Statistics
Pi
313.746828
Theta
169.425375
Tajima's D
2.314537
CLR
0.429145
CSRT
0.926103694815259
Interpretation
Uncertain
