Gene
KWMTBOMO03435
Pre Gene Modal
BGIBMGA006367
Annotation
PREDICTED:_protein_unc-80_homolog_[Bombyx_mori]
Full name
Protein unc-80 homolog
Location in the cell
Nuclear Reliability : 1.855
Sequence
CDS
ATGAATCGAGAAAATCTACTAGAACTGAAGGAAGCTTGTAAGTCGTTCGAAAAAGTACTAGTTCAAAATTTGAGATTAGGTCTCTCAAGCAGTCTCACTGAGGCTATTGAATCAATACCACGATGGCGCCTCATACAAGCAGCACTGCCTCATGTCATGCATTCCTTGGCATCAGTGCTTCACACAAGAGTAAAGGACAATATGCAGTCACTTGGAAATGTGGAAACAAAGCTGTTGTATACTCTTCACTGGATCCTTCTCGATGCTTCCGATGAATGTGCTGACAATGACTATGAAAATGGCGTATTTTATTCAAACCCATTTCACTATTTGTTTTCAGTACCAACAATGACGCTATTTGTATTTCTGTTTGCACCACTTTCGCACCACCTGAAAGAATCAGACTTCACCAATAACTATCGCTTAGAAAATGGTCTAAAGATGTGGCAAGCGATGTGGGAATTTCGGCATCCTGATGCGCCTTGTTTTACAACACTTGTGAAACCAAAACCGAAGCCTTTGGGTGGAAAATATTCGAAAAGGCCACAACACAGCGACGTATTTCTTGGAAGGAAATTTTCAAAAGAAGAAGGCACGGTCAATGGTAGTCCACCAAGTCAGAATGTATCCCTATCACAGTCGGATGTGCCATCCAAACAAGCCAGCACTGAGGAGGAGCCGTGGCTATCATCACCTAAAGAAACTATTTTTCCAGAAACAATACCCGAAGAGAGTTCGAGCACCGAAGAAGAACATGTGTTTATATACCGACTTCCCTCCGAACAAAATTTTGGTGGTTCCGTGAAAGAGGCCAGTCGTTACCAGGGATAA
Protein
MNRENLLELKEACKSFEKVLVQNLRLGLSSSLTEAIESIPRWRLIQAALPHVMHSLASVLHTRVKDNMQSLGNVETKLLYTLHWILLDASDECADNDYENGVFYSNPFHYLFSVPTMTLFVFLFAPLSHHLKESDFTNNYRLENGLKMWQAMWEFRHPDAPCFTTLVKPKPKPLGGKYSKRPQHSDVFLGRKFSKEEGTVNGSPPSQNVSLSQSDVPSKQASTEEEPWLSSPKETIFPETIPEESSSTEEEHVFIYRLPSEQNFGGSVKEASRYQG
Summary
Description
Component of the na (narrow abdomen) sodium channel complex. In the circadian clock neurons it functions with na and unc79 to promote circadian rhythmicity.
Subunit
Interacts with unc79 and na. Can interact with unc79 independently of na.
Similarity
Belongs to the unc-80 family.
Keywords
Complete proteome
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Protein unc-80 homolog
Uniprot
H9JA22
A0A194RNK8
A0A194PXL4
A0A2H1V856
A0A212F3A3
A0A2A4JD08
+ More
A0A2A4JBI4 A0A2A4JBF5 A0A0L7L9D2 A0A2J7Q8G6 D7EIC7 A0A139WNP9 A0A139WNA1 A0A139WNA3 U4U2Y5 N6TSD9 A0A1B6MM87 A0A2P8ZPM7 A0A088AJQ7 E0VJG7 T1I3F0 A0A182TN28 A0A182TBK4 A0A088AJQ8 A0A034WR41 A0A1B0CSU9 A0A182WSD6 A0A182HL87 F5HML1 Q8T5I0 A0A2R7WR13 A0A182R661 A0A182F7L8 A0A182VZ79 A0A182MTD3 A0A182PGS7 A0A158NMS2 A0A182L9M3 E2BEX5 A0A182JHE2 A0A0L0C2W3 B4QY27 A0A0R1E518 B4NAR2 B4JRX7 B3LX38 A0A1J1HNV7 W5J732 A0A1A9Z7B3 A0A0R1EA21 B4PRB5 A0A0R1E6A3 A0A0R1E4Z3 A0A0R1E9G6 A0A0R1E4Y7 A0A0R1E4Z1 A0A0R1E5K1 A0A0R1E5F2 A0A0R1EA26 A0A182UPQ0 B3P5Y9 A0A0R1E847 B4IGU7 B4M624 A0A195DBP2 A0A0B4KI46 A0A0B4KH56 A0A1W4URK2 A0A1W4UCM6 A0A1W4URK7 A0A1W4UPZ1 A0A1W4UPP8 A0A0B4KHR1 A0A1W4UCL6 A0A1W4UD65 A0A1W4UPQ4 A0A1W4UCM1 A0A1W4UPZ8 Q9VB11 A0A1W4UPQ8 A0A0B4KHA4 E2ALP1 A0A151I0X9 A0A0L7R4E4 B4KB92 A0A151X159 A0A195C7R5 Q29BG7 B4GNX6 A0A1A9VJ27 A0A151JVR1 A0A2P6KQM2 A0A0R3NF38 A0A0R3NF00 A0A0R3NKF9 A0A0R3NFR7 A0A182N426 A0A0R3NLI1 A0A0R3NFD4 A0A0R3NKR3
A0A2A4JBI4 A0A2A4JBF5 A0A0L7L9D2 A0A2J7Q8G6 D7EIC7 A0A139WNP9 A0A139WNA1 A0A139WNA3 U4U2Y5 N6TSD9 A0A1B6MM87 A0A2P8ZPM7 A0A088AJQ7 E0VJG7 T1I3F0 A0A182TN28 A0A182TBK4 A0A088AJQ8 A0A034WR41 A0A1B0CSU9 A0A182WSD6 A0A182HL87 F5HML1 Q8T5I0 A0A2R7WR13 A0A182R661 A0A182F7L8 A0A182VZ79 A0A182MTD3 A0A182PGS7 A0A158NMS2 A0A182L9M3 E2BEX5 A0A182JHE2 A0A0L0C2W3 B4QY27 A0A0R1E518 B4NAR2 B4JRX7 B3LX38 A0A1J1HNV7 W5J732 A0A1A9Z7B3 A0A0R1EA21 B4PRB5 A0A0R1E6A3 A0A0R1E4Z3 A0A0R1E9G6 A0A0R1E4Y7 A0A0R1E4Z1 A0A0R1E5K1 A0A0R1E5F2 A0A0R1EA26 A0A182UPQ0 B3P5Y9 A0A0R1E847 B4IGU7 B4M624 A0A195DBP2 A0A0B4KI46 A0A0B4KH56 A0A1W4URK2 A0A1W4UCM6 A0A1W4URK7 A0A1W4UPZ1 A0A1W4UPP8 A0A0B4KHR1 A0A1W4UCL6 A0A1W4UD65 A0A1W4UPQ4 A0A1W4UCM1 A0A1W4UPZ8 Q9VB11 A0A1W4UPQ8 A0A0B4KHA4 E2ALP1 A0A151I0X9 A0A0L7R4E4 B4KB92 A0A151X159 A0A195C7R5 Q29BG7 B4GNX6 A0A1A9VJ27 A0A151JVR1 A0A2P6KQM2 A0A0R3NF38 A0A0R3NF00 A0A0R3NKF9 A0A0R3NFR7 A0A182N426 A0A0R3NLI1 A0A0R3NFD4 A0A0R3NKR3
Pubmed
19121390
26354079
22118469
26227816
18362917
19820115
+ More
23537049 29403074 20566863 25348373 12364791 12060762 14747013 17210077 21347285 20966253 20798317 26108605 17994087 17550304 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17350263 24223770 15632085
23537049 29403074 20566863 25348373 12364791 12060762 14747013 17210077 21347285 20966253 20798317 26108605 17994087 17550304 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17350263 24223770 15632085
EMBL
BABH01009293
BABH01009294
BABH01009295
BABH01009296
BABH01009297
BABH01009298
+ More
KQ459896 KPJ19423.1 KQ459589 KPI97499.1 ODYU01001175 SOQ37033.1 AGBW02010586 OWR48218.1 NWSH01002013 PCG69434.1 PCG69435.1 PCG69437.1 JTDY01002227 KOB71851.1 NEVH01016955 PNF24872.1 KQ971310 EFA11788.2 KYB29507.1 KYB29508.1 KYB29509.1 KB631850 ERL86703.1 APGK01057001 APGK01057002 APGK01057003 KB741277 ENN71241.1 GEBQ01002919 JAT37058.1 PYGN01000001 PSN58458.1 DS235222 EEB13523.1 ACPB03010432 GAKP01002297 JAC56655.1 AJWK01026635 AJWK01026636 APCN01000884 AAAB01008987 EGK97533.1 AJ439353 CAD27932.1 EAA00911.2 KK855320 PTY21998.1 AXCM01003048 ADTU01020713 ADTU01020714 GL447885 EFN85740.1 JRES01000972 KNC26665.1 CM000364 EDX14675.1 CM000160 KRK04285.1 CH964232 EDW80876.2 CH916373 EDV94517.1 CH902617 EDV43877.2 CVRI01000006 CRK87913.1 ADMH02001961 ETN60267.1 KRK04292.1 EDW98479.2 KRK04286.1 KRK04288.1 KRK04289.1 KRK04283.1 KRK04284.1 KRK04287.1 KRK04293.1 KRK04290.1 CH954182 EDV53389.1 KRK04291.1 CH480837 EDW49065.1 CH940652 EDW59100.2 KQ981010 KYN10340.1 AE014297 AGB96405.1 AGB96403.1 AGB96404.1 AGB96402.1 GL440609 EFN65650.1 KQ976593 KYM79683.1 KQ414657 KOC65755.1 CH933806 EDW16820.2 KQ982588 KYQ54096.1 KQ978231 KYM96206.1 CM000070 EAL27031.4 CH479186 EDW38859.1 KQ981689 KYN37900.1 MWRG01006891 PRD28608.1 KRS99734.1 KRS99736.1 KRS99740.1 KRS99739.1 KRS99733.1 KRS99738.1 KRS99730.1
KQ459896 KPJ19423.1 KQ459589 KPI97499.1 ODYU01001175 SOQ37033.1 AGBW02010586 OWR48218.1 NWSH01002013 PCG69434.1 PCG69435.1 PCG69437.1 JTDY01002227 KOB71851.1 NEVH01016955 PNF24872.1 KQ971310 EFA11788.2 KYB29507.1 KYB29508.1 KYB29509.1 KB631850 ERL86703.1 APGK01057001 APGK01057002 APGK01057003 KB741277 ENN71241.1 GEBQ01002919 JAT37058.1 PYGN01000001 PSN58458.1 DS235222 EEB13523.1 ACPB03010432 GAKP01002297 JAC56655.1 AJWK01026635 AJWK01026636 APCN01000884 AAAB01008987 EGK97533.1 AJ439353 CAD27932.1 EAA00911.2 KK855320 PTY21998.1 AXCM01003048 ADTU01020713 ADTU01020714 GL447885 EFN85740.1 JRES01000972 KNC26665.1 CM000364 EDX14675.1 CM000160 KRK04285.1 CH964232 EDW80876.2 CH916373 EDV94517.1 CH902617 EDV43877.2 CVRI01000006 CRK87913.1 ADMH02001961 ETN60267.1 KRK04292.1 EDW98479.2 KRK04286.1 KRK04288.1 KRK04289.1 KRK04283.1 KRK04284.1 KRK04287.1 KRK04293.1 KRK04290.1 CH954182 EDV53389.1 KRK04291.1 CH480837 EDW49065.1 CH940652 EDW59100.2 KQ981010 KYN10340.1 AE014297 AGB96405.1 AGB96403.1 AGB96404.1 AGB96402.1 GL440609 EFN65650.1 KQ976593 KYM79683.1 KQ414657 KOC65755.1 CH933806 EDW16820.2 KQ982588 KYQ54096.1 KQ978231 KYM96206.1 CM000070 EAL27031.4 CH479186 EDW38859.1 KQ981689 KYN37900.1 MWRG01006891 PRD28608.1 KRS99734.1 KRS99736.1 KRS99740.1 KRS99739.1 KRS99733.1 KRS99738.1 KRS99730.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000218220
UP000037510
+ More
UP000235965 UP000007266 UP000030742 UP000019118 UP000245037 UP000005203 UP000009046 UP000015103 UP000075902 UP000075901 UP000092461 UP000076407 UP000075840 UP000007062 UP000075900 UP000069272 UP000075920 UP000075883 UP000075885 UP000005205 UP000075882 UP000008237 UP000075880 UP000037069 UP000000304 UP000002282 UP000007798 UP000001070 UP000007801 UP000183832 UP000000673 UP000092445 UP000075903 UP000008711 UP000001292 UP000008792 UP000078492 UP000000803 UP000192221 UP000000311 UP000078540 UP000053825 UP000009192 UP000075809 UP000078542 UP000001819 UP000008744 UP000078200 UP000078541 UP000075884
UP000235965 UP000007266 UP000030742 UP000019118 UP000245037 UP000005203 UP000009046 UP000015103 UP000075902 UP000075901 UP000092461 UP000076407 UP000075840 UP000007062 UP000075900 UP000069272 UP000075920 UP000075883 UP000075885 UP000005205 UP000075882 UP000008237 UP000075880 UP000037069 UP000000304 UP000002282 UP000007798 UP000001070 UP000007801 UP000183832 UP000000673 UP000092445 UP000075903 UP000008711 UP000001292 UP000008792 UP000078492 UP000000803 UP000192221 UP000000311 UP000078540 UP000053825 UP000009192 UP000075809 UP000078542 UP000001819 UP000008744 UP000078200 UP000078541 UP000075884
Pfam
Interpro
ProteinModelPortal
H9JA22
A0A194RNK8
A0A194PXL4
A0A2H1V856
A0A212F3A3
A0A2A4JD08
+ More
A0A2A4JBI4 A0A2A4JBF5 A0A0L7L9D2 A0A2J7Q8G6 D7EIC7 A0A139WNP9 A0A139WNA1 A0A139WNA3 U4U2Y5 N6TSD9 A0A1B6MM87 A0A2P8ZPM7 A0A088AJQ7 E0VJG7 T1I3F0 A0A182TN28 A0A182TBK4 A0A088AJQ8 A0A034WR41 A0A1B0CSU9 A0A182WSD6 A0A182HL87 F5HML1 Q8T5I0 A0A2R7WR13 A0A182R661 A0A182F7L8 A0A182VZ79 A0A182MTD3 A0A182PGS7 A0A158NMS2 A0A182L9M3 E2BEX5 A0A182JHE2 A0A0L0C2W3 B4QY27 A0A0R1E518 B4NAR2 B4JRX7 B3LX38 A0A1J1HNV7 W5J732 A0A1A9Z7B3 A0A0R1EA21 B4PRB5 A0A0R1E6A3 A0A0R1E4Z3 A0A0R1E9G6 A0A0R1E4Y7 A0A0R1E4Z1 A0A0R1E5K1 A0A0R1E5F2 A0A0R1EA26 A0A182UPQ0 B3P5Y9 A0A0R1E847 B4IGU7 B4M624 A0A195DBP2 A0A0B4KI46 A0A0B4KH56 A0A1W4URK2 A0A1W4UCM6 A0A1W4URK7 A0A1W4UPZ1 A0A1W4UPP8 A0A0B4KHR1 A0A1W4UCL6 A0A1W4UD65 A0A1W4UPQ4 A0A1W4UCM1 A0A1W4UPZ8 Q9VB11 A0A1W4UPQ8 A0A0B4KHA4 E2ALP1 A0A151I0X9 A0A0L7R4E4 B4KB92 A0A151X159 A0A195C7R5 Q29BG7 B4GNX6 A0A1A9VJ27 A0A151JVR1 A0A2P6KQM2 A0A0R3NF38 A0A0R3NF00 A0A0R3NKF9 A0A0R3NFR7 A0A182N426 A0A0R3NLI1 A0A0R3NFD4 A0A0R3NKR3
A0A2A4JBI4 A0A2A4JBF5 A0A0L7L9D2 A0A2J7Q8G6 D7EIC7 A0A139WNP9 A0A139WNA1 A0A139WNA3 U4U2Y5 N6TSD9 A0A1B6MM87 A0A2P8ZPM7 A0A088AJQ7 E0VJG7 T1I3F0 A0A182TN28 A0A182TBK4 A0A088AJQ8 A0A034WR41 A0A1B0CSU9 A0A182WSD6 A0A182HL87 F5HML1 Q8T5I0 A0A2R7WR13 A0A182R661 A0A182F7L8 A0A182VZ79 A0A182MTD3 A0A182PGS7 A0A158NMS2 A0A182L9M3 E2BEX5 A0A182JHE2 A0A0L0C2W3 B4QY27 A0A0R1E518 B4NAR2 B4JRX7 B3LX38 A0A1J1HNV7 W5J732 A0A1A9Z7B3 A0A0R1EA21 B4PRB5 A0A0R1E6A3 A0A0R1E4Z3 A0A0R1E9G6 A0A0R1E4Y7 A0A0R1E4Z1 A0A0R1E5K1 A0A0R1E5F2 A0A0R1EA26 A0A182UPQ0 B3P5Y9 A0A0R1E847 B4IGU7 B4M624 A0A195DBP2 A0A0B4KI46 A0A0B4KH56 A0A1W4URK2 A0A1W4UCM6 A0A1W4URK7 A0A1W4UPZ1 A0A1W4UPP8 A0A0B4KHR1 A0A1W4UCL6 A0A1W4UD65 A0A1W4UPQ4 A0A1W4UCM1 A0A1W4UPZ8 Q9VB11 A0A1W4UPQ8 A0A0B4KHA4 E2ALP1 A0A151I0X9 A0A0L7R4E4 B4KB92 A0A151X159 A0A195C7R5 Q29BG7 B4GNX6 A0A1A9VJ27 A0A151JVR1 A0A2P6KQM2 A0A0R3NF38 A0A0R3NF00 A0A0R3NKF9 A0A0R3NFR7 A0A182N426 A0A0R3NLI1 A0A0R3NFD4 A0A0R3NKR3
Ontologies
GO
Topology
Subcellular location
Membrane
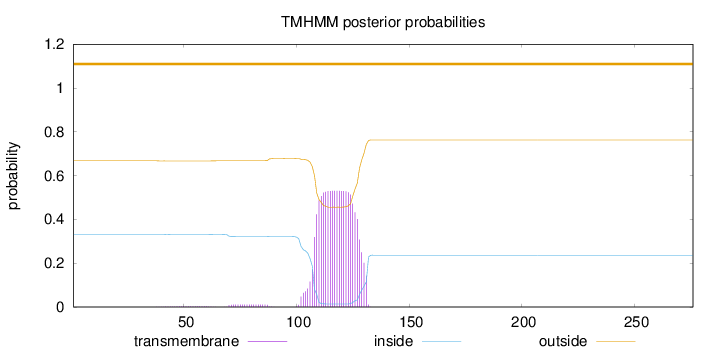
Length:
276
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
11.62142
Exp number, first 60 AAs:
0.0491
Total prob of N-in:
0.33258
outside
1 - 276
Population Genetic Test Statistics
Pi
176.237832
Theta
145.418873
Tajima's D
0.117561
CLR
0.051307
CSRT
0.409729513524324
Interpretation
Uncertain
