Gene
KWMTBOMO03430 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006507
Annotation
glutamate_dehydrogenase_[Bombyx_mori]
Full name
Glutamate dehydrogenase
+ More
Glutamate dehydrogenase, mitochondrial
Glutamate dehydrogenase, mitochondrial
Location in the cell
Cytoplasmic Reliability : 2.33
Sequence
CDS
ATGCTGCATCTCAAGAATATCGCCAAGTCAGTGGTTCCGCCGCTCAAGAATTCTGTTCAAAATGAAGCCCTCAATACAATGTTCCGAATCATACCAGCTGGAGTGAATGTCTGCTGCCGCACATACGCTAGTCATGAGATTCCAGATAAGCTCAAAGATATTCCTACAAGTGCGAATCCGAAGTTCTTCCACATGGTAGAATATTTTTTCCACCGAGCCTGTCAAGTTGTCGAAGACAAGCTTGTTGAAGATTTGAAGTCAAGGACACCCATTGAAGAGAAGAAAAAGAAAGTAGCCGGTATTCTAAAACTTATGGAACCATGCGATCACATTCTTGAGATTCAATTTCCTCTGAGGCGCGATTCTGGCGATTACGAAATGATATTAGGCTATCGCGCACAACATTCCACACACAGGACTCCAACCAAAGGAGGTATTCGATTCTCAACGGACGTAACCAGAGATGAAGTTAAGGCGTTATCAGCTTTGATGACCTTCAAGTGCGCGTGCGTGGACGTGCCTTTCGGCGGTGCTAAGGCCGGTATCAAGATCAATCCCAAAGAATACTCCGAGCATGAACTGGAAAAGATCACTCGTCGTTTCACCCTTGAACTTGCCAAAAAAGGATTCATTGGGCCTGGCGTGGATGTCCCCGCTCCTGACATGGGTACCGGCGAACGAGAAATGTCTTGGATCGCCGATACTTATGCGAAGACCGTCGGTTTTCAAGACATCAACGCTCACGCCTGCGTCACTGGCAAACCTATTAACCAGGGTGGCATCCACGGCAGAGTTTCAGCCACGGGCAGGGGCGTATTCCACGGCTTGGAGAACTTCATCAACGAAGCCAACTACATGAGCATGATCGGTACAACCCCCGGTTGGGGTGGCAAGACGTTCATCGTCCAAGGTTTCGGTAACGTGGGACTCCACACTTGCCGCTACCTCGTCCGCGCCGGCGCCACTTGCATCGGAGTTATCGAGCACGACGGCTCCATTTACAACCCTGATGGCATCAACCCTAAGGCCTTGGAGGACTACAGAATCGAGAACGGTACGGTAGTCGGTTTCCCCGGCGCTAAGGCCTACGAAGGCGAGAACATGCTTTACGAGAAGTGCGACATTCTTGTACCCGCCGCCATCGAACAGGTCATAAACAAGGACAACGCTCACAGGATCCAAGCTAAGATCATTGCGGAGGCCGCCAACGGTCCCACCACACCTGCTGCAGACAAGATCCTCATCGATCGCAACATTCTCGTGATCCCCGACCTCTACATCAACGCTGGTGGTGTCACCGTCTCATTCTTCGAGTGGCTCAAGAACCTCAATCACGTGTCTTACGGACGTCTGACATTCAAATACGAGAGGGAATCTAACTACCATCTGCTGGAATCGGTCCAAGAGTCTCTCGAGCGGCGGTTCGGTCGCGTGGGAGGCCGCATCCCCGTCACTCCCTCAGAGTCCTTCCAGAAGAGAATCTCCGGCGCCTCCGAGAAGGACATCGTGCACTCCGGACTCGACTACACCATGGAGAGATCCGCTAGGGCCATCATGAAGACAGCCATGAGGTTCAACCTCGGTTTAGATCTGAGGACAGCCGCGTATGCGAACTCCATCGAAAAGATATTCACCACTGTCGAGTTAACGTTTGAACTTCCTGATAGTGCCGAAGAATGTTTTTATCAAACAATGGACAAGGATACGGCTGCTTCATTGGAGTACCAAGTAATTTCTGGAGGACAGTATGACGTAAGCGTAAAAATAGAAGCTCCTAACAGACAAATTATATATCAGCAAGAGAAGATGCAATATGATTCACACCAGTTCACCGCTCTGGCGTCTGGTGATTACAAAGTGTGTTTTAGTAATGAGTTTAGTACATTTTCACATAAACTTGTGTATATGGAACTAAATGTTGGACCGGAGCAGCCTCTTCCTGGTGTAGGAGATCATGCAACTGTTCTTACACAGTTGGAGACATCTGCCGAAGAAATTCATTCTGCACTCAACAGAATCATTGATCATCAAACTCATCATAGATTAAGAGAGGCTCAAGGACGTAAAAGAGCTGAAGATCTGAATGAAAAAGTATTTTGGTGGTCTATGGGTGAAACCTTGGCTATTGTTTGTGTTGCATTTACACAAGTTATGATACTAAAGAATTTCTTTAGTGACAAGCCTACTCTATACAACCGATTGTAA
Protein
MLHLKNIAKSVVPPLKNSVQNEALNTMFRIIPAGVNVCCRTYASHEIPDKLKDIPTSANPKFFHMVEYFFHRACQVVEDKLVEDLKSRTPIEEKKKKVAGILKLMEPCDHILEIQFPLRRDSGDYEMILGYRAQHSTHRTPTKGGIRFSTDVTRDEVKALSALMTFKCACVDVPFGGAKAGIKINPKEYSEHELEKITRRFTLELAKKGFIGPGVDVPAPDMGTGEREMSWIADTYAKTVGFQDINAHACVTGKPINQGGIHGRVSATGRGVFHGLENFINEANYMSMIGTTPGWGGKTFIVQGFGNVGLHTCRYLVRAGATCIGVIEHDGSIYNPDGINPKALEDYRIENGTVVGFPGAKAYEGENMLYEKCDILVPAAIEQVINKDNAHRIQAKIIAEAANGPTTPAADKILIDRNILVIPDLYINAGGVTVSFFEWLKNLNHVSYGRLTFKYERESNYHLLESVQESLERRFGRVGGRIPVTPSESFQKRISGASEKDIVHSGLDYTMERSARAIMKTAMRFNLGLDLRTAAYANSIEKIFTTVELTFELPDSAEECFYQTMDKDTAASLEYQVISGGQYDVSVKIEAPNRQIIYQQEKMQYDSHQFTALASGDYKVCFSNEFSTFSHKLVYMELNVGPEQPLPGVGDHATVLTQLETSAEEIHSALNRIIDHQTHHRLREAQGRKRAEDLNEKVFWWSMGETLAIVCVAFTQVMILKNFFSDKPTLYNRL
Summary
Catalytic Activity
H2O + L-glutamate + NAD(+) = 2-oxoglutarate + H(+) + NADH + NH4(+)
H2O + L-glutamate + NADP(+) = 2-oxoglutarate + H(+) + NADPH + NH4(+)
H2O + L-glutamate + NADP(+) = 2-oxoglutarate + H(+) + NADPH + NH4(+)
Subunit
Homohexamer.
Miscellaneous
ADP can occupy the NADH binding site and activate the enzyme.
Similarity
Belongs to the Glu/Leu/Phe/Val dehydrogenases family.
Keywords
Alternative splicing
ATP-binding
Complete proteome
GTP-binding
Mitochondrion
NAD
Nucleotide-binding
Oxidoreductase
Reference proteome
Transit peptide
Feature
chain Glutamate dehydrogenase, mitochondrial
splice variant In isoform C.
splice variant In isoform C.
Uniprot
Q2F5R4
A0A2A4JCS9
A0A0G3VFZ9
A0A194Q273
S4PE01
I4DQZ0
+ More
A0A194RNK3 I4DJ42 A0A0K8TT23 E2C3C7 E9IPG3 A0A310SFZ1 A0A151X7U3 A0A2A3EPX9 A0A087ZS68 E2A6U2 A0A2J7PVJ1 A0A195FTW7 A0A154PIZ3 A0A158NNF9 W8BYK3 A0A1J1IUJ7 A0A068TJS2 A0A026WII5 A0A034VPU7 A0A0A1XFV2 A0A1I8NSV1 A0A1B6GVV3 A0A336MGF4 T1PBD9 A0A232F5M0 K7IYA3 A0A0M9AAI4 A0A195C1R0 A0A1L8EEM7 A0A158NNF3 A0A1B6EYJ5 A0A1L8E2M4 P54385-2 A0A0P8ZYJ0 A0A0Q9WUT7 A0A0R1EBF8 A0A182QQS6 A0A1W4U8M3 A0A1J1IWE9 A0A034VKL6 A0A2M4CUS3 A0A0Q9WR91 A0A3B0KCM3 A0A1I8NSU8 I5ANX6 A0A2M4CT81 A0A0C9Q076 A0A0Q9X1Y5 A0A0A1X040 A0A1A9Y0I5 A0A1B0B2Z8 T1P987 A0A1A9W5Q9 D3TQR2 A0A1A9ZSU9 A0A0K8UFK5 A0A2M3ZZI4 A0A336KTS7 A0A182R8H6 A0A1L8EER7 F5HLL7 A0A2M3YYJ1 B3M317 A0A0L0BRA3 A0A182XF30 A0A0M3QYL6 B4N9E8 B4PL48 B3P8B7 P54385 A0A1W4UJV6 B4QSD4 U5EZ79 A0A3B0JQU1 B4HG10 Q298M7 B4G4C1 A0A182LW14 B4K740 B4M0C5 A0A182PG80 A0A023ETW6 A0A0L7RDB5 B4JRQ0 Q2KQ98 A0A2M3ZZC8 A0A182IW02 A0A182V234 A0A182UC31 A0A182HVJ8 A0A182YJQ2
A0A194RNK3 I4DJ42 A0A0K8TT23 E2C3C7 E9IPG3 A0A310SFZ1 A0A151X7U3 A0A2A3EPX9 A0A087ZS68 E2A6U2 A0A2J7PVJ1 A0A195FTW7 A0A154PIZ3 A0A158NNF9 W8BYK3 A0A1J1IUJ7 A0A068TJS2 A0A026WII5 A0A034VPU7 A0A0A1XFV2 A0A1I8NSV1 A0A1B6GVV3 A0A336MGF4 T1PBD9 A0A232F5M0 K7IYA3 A0A0M9AAI4 A0A195C1R0 A0A1L8EEM7 A0A158NNF3 A0A1B6EYJ5 A0A1L8E2M4 P54385-2 A0A0P8ZYJ0 A0A0Q9WUT7 A0A0R1EBF8 A0A182QQS6 A0A1W4U8M3 A0A1J1IWE9 A0A034VKL6 A0A2M4CUS3 A0A0Q9WR91 A0A3B0KCM3 A0A1I8NSU8 I5ANX6 A0A2M4CT81 A0A0C9Q076 A0A0Q9X1Y5 A0A0A1X040 A0A1A9Y0I5 A0A1B0B2Z8 T1P987 A0A1A9W5Q9 D3TQR2 A0A1A9ZSU9 A0A0K8UFK5 A0A2M3ZZI4 A0A336KTS7 A0A182R8H6 A0A1L8EER7 F5HLL7 A0A2M3YYJ1 B3M317 A0A0L0BRA3 A0A182XF30 A0A0M3QYL6 B4N9E8 B4PL48 B3P8B7 P54385 A0A1W4UJV6 B4QSD4 U5EZ79 A0A3B0JQU1 B4HG10 Q298M7 B4G4C1 A0A182LW14 B4K740 B4M0C5 A0A182PG80 A0A023ETW6 A0A0L7RDB5 B4JRQ0 Q2KQ98 A0A2M3ZZC8 A0A182IW02 A0A182V234 A0A182UC31 A0A182HVJ8 A0A182YJQ2
EC Number
1.4.1.3
Pubmed
19121390
26354079
23622113
22651552
26369729
20798317
+ More
21282665 21347285 24495485 24508170 25348373 25830018 25315136 28648823 20075255 10992165 10731132 12537572 12537569 6810872 17994087 17550304 15632085 20353571 12364791 14747013 17210077 26108605 24945155 26483478 15804581 17510324 25244985
21282665 21347285 24495485 24508170 25348373 25830018 25315136 28648823 20075255 10992165 10731132 12537572 12537569 6810872 17994087 17550304 15632085 20353571 12364791 14747013 17210077 26108605 24945155 26483478 15804581 17510324 25244985
EMBL
BABH01009287
BABH01009288
BABH01009289
BABH01009290
DQ311359
ABD36303.1
+ More
NWSH01001856 PCG69907.1 KP657639 RSAL01000283 AKL78864.1 RVE43001.1 KQ459589 KPI97505.1 GAIX01003416 JAA89144.1 AK404850 BAM20330.1 KQ459896 KPJ19418.1 AK401310 BAM17932.1 GDAI01000089 JAI17514.1 GL452320 EFN77465.1 GL764503 EFZ17554.1 KQ760382 OAD60710.1 KQ982431 KYQ56433.1 KZ288204 PBC33189.1 GL437203 EFN70808.1 NEVH01020956 PNF20354.1 KQ981268 KYN43903.1 KQ434931 KZC11856.1 ADTU01002703 ADTU01002704 ADTU01002705 ADTU01002706 ADTU01002707 ADTU01002708 ADTU01002709 ADTU01002710 ADTU01002711 ADTU01002712 GAMC01008174 JAB98381.1 CVRI01000060 CRL03893.1 HG965788 CDO39384.1 KK107234 EZA54914.1 GAKP01015117 JAC43835.1 GBXI01004909 JAD09383.1 GECZ01003209 GECZ01000641 JAS66560.1 JAS69128.1 UFQS01000897 UFQT01000897 SSX07543.1 SSX27883.1 KA645293 AFP59922.1 NNAY01000875 OXU26104.1 KQ435714 KOX79199.1 KQ978344 KYM94782.1 GFDG01001617 JAV17182.1 GECZ01026752 GECZ01010743 JAS43017.1 JAS59026.1 GFDF01001150 JAV12934.1 Y11314 Z29062 AE014297 AY061323 BT001501 CH902617 KPU79715.1 CH964232 KRF99304.1 CM000160 KRK04628.1 AXCN02000334 CRL03892.1 GAKP01015116 JAC43836.1 GGFL01004733 MBW68911.1 CH940650 KRF83230.1 OUUW01000007 SPP82761.1 CM000070 EIM52661.1 GGFL01004352 MBW68530.1 GBYB01007233 JAG77000.1 CH933806 KRG02059.1 GBXI01009825 JAD04467.1 JXJN01007741 JXJN01007742 JXJN01007743 KA645292 AFP59921.1 EZ423764 ADD20040.1 GDHF01026885 JAI25429.1 GGFK01000665 MBW33986.1 SSX07542.1 SSX27882.1 GFDG01001619 JAV17180.1 AAAB01008898 EGK97150.1 GGFM01000592 MBW21343.1 EDV42417.1 JRES01001482 KNC22546.1 CP012526 ALC47893.1 EDW80581.1 EDW99033.1 CH954182 EDV53941.1 CM000364 EDX14133.1 GANO01001279 JAB58592.1 SPP82762.1 CH480815 EDW43403.1 EAL27928.2 CH479179 EDW24469.1 AXCM01000759 EDW16353.1 EDW67287.1 JXUM01016134 JXUM01016135 GAPW01000805 KQ560450 JAC12793.1 KXJ82361.1 KQ414615 KOC68746.1 CH916373 EDV94440.1 AY623405 CH477664 AAV31914.1 EAT37557.1 EAT37559.1 GGFK01000605 MBW33926.1 APCN01005788
NWSH01001856 PCG69907.1 KP657639 RSAL01000283 AKL78864.1 RVE43001.1 KQ459589 KPI97505.1 GAIX01003416 JAA89144.1 AK404850 BAM20330.1 KQ459896 KPJ19418.1 AK401310 BAM17932.1 GDAI01000089 JAI17514.1 GL452320 EFN77465.1 GL764503 EFZ17554.1 KQ760382 OAD60710.1 KQ982431 KYQ56433.1 KZ288204 PBC33189.1 GL437203 EFN70808.1 NEVH01020956 PNF20354.1 KQ981268 KYN43903.1 KQ434931 KZC11856.1 ADTU01002703 ADTU01002704 ADTU01002705 ADTU01002706 ADTU01002707 ADTU01002708 ADTU01002709 ADTU01002710 ADTU01002711 ADTU01002712 GAMC01008174 JAB98381.1 CVRI01000060 CRL03893.1 HG965788 CDO39384.1 KK107234 EZA54914.1 GAKP01015117 JAC43835.1 GBXI01004909 JAD09383.1 GECZ01003209 GECZ01000641 JAS66560.1 JAS69128.1 UFQS01000897 UFQT01000897 SSX07543.1 SSX27883.1 KA645293 AFP59922.1 NNAY01000875 OXU26104.1 KQ435714 KOX79199.1 KQ978344 KYM94782.1 GFDG01001617 JAV17182.1 GECZ01026752 GECZ01010743 JAS43017.1 JAS59026.1 GFDF01001150 JAV12934.1 Y11314 Z29062 AE014297 AY061323 BT001501 CH902617 KPU79715.1 CH964232 KRF99304.1 CM000160 KRK04628.1 AXCN02000334 CRL03892.1 GAKP01015116 JAC43836.1 GGFL01004733 MBW68911.1 CH940650 KRF83230.1 OUUW01000007 SPP82761.1 CM000070 EIM52661.1 GGFL01004352 MBW68530.1 GBYB01007233 JAG77000.1 CH933806 KRG02059.1 GBXI01009825 JAD04467.1 JXJN01007741 JXJN01007742 JXJN01007743 KA645292 AFP59921.1 EZ423764 ADD20040.1 GDHF01026885 JAI25429.1 GGFK01000665 MBW33986.1 SSX07542.1 SSX27882.1 GFDG01001619 JAV17180.1 AAAB01008898 EGK97150.1 GGFM01000592 MBW21343.1 EDV42417.1 JRES01001482 KNC22546.1 CP012526 ALC47893.1 EDW80581.1 EDW99033.1 CH954182 EDV53941.1 CM000364 EDX14133.1 GANO01001279 JAB58592.1 SPP82762.1 CH480815 EDW43403.1 EAL27928.2 CH479179 EDW24469.1 AXCM01000759 EDW16353.1 EDW67287.1 JXUM01016134 JXUM01016135 GAPW01000805 KQ560450 JAC12793.1 KXJ82361.1 KQ414615 KOC68746.1 CH916373 EDV94440.1 AY623405 CH477664 AAV31914.1 EAT37557.1 EAT37559.1 GGFK01000605 MBW33926.1 APCN01005788
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000008237
+ More
UP000075809 UP000242457 UP000005203 UP000000311 UP000235965 UP000078541 UP000076502 UP000005205 UP000183832 UP000053097 UP000095300 UP000095301 UP000215335 UP000002358 UP000053105 UP000078542 UP000000803 UP000007801 UP000007798 UP000002282 UP000075886 UP000192221 UP000008792 UP000268350 UP000001819 UP000009192 UP000092443 UP000092460 UP000091820 UP000092445 UP000075900 UP000007062 UP000037069 UP000076407 UP000092553 UP000008711 UP000000304 UP000001292 UP000008744 UP000075883 UP000075885 UP000069940 UP000249989 UP000053825 UP000001070 UP000008820 UP000075880 UP000075903 UP000075902 UP000075840 UP000076408
UP000075809 UP000242457 UP000005203 UP000000311 UP000235965 UP000078541 UP000076502 UP000005205 UP000183832 UP000053097 UP000095300 UP000095301 UP000215335 UP000002358 UP000053105 UP000078542 UP000000803 UP000007801 UP000007798 UP000002282 UP000075886 UP000192221 UP000008792 UP000268350 UP000001819 UP000009192 UP000092443 UP000092460 UP000091820 UP000092445 UP000075900 UP000007062 UP000037069 UP000076407 UP000092553 UP000008711 UP000000304 UP000001292 UP000008744 UP000075883 UP000075885 UP000069940 UP000249989 UP000053825 UP000001070 UP000008820 UP000075880 UP000075903 UP000075902 UP000075840 UP000076408
PRIDE
Interpro
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
Q2F5R4
A0A2A4JCS9
A0A0G3VFZ9
A0A194Q273
S4PE01
I4DQZ0
+ More
A0A194RNK3 I4DJ42 A0A0K8TT23 E2C3C7 E9IPG3 A0A310SFZ1 A0A151X7U3 A0A2A3EPX9 A0A087ZS68 E2A6U2 A0A2J7PVJ1 A0A195FTW7 A0A154PIZ3 A0A158NNF9 W8BYK3 A0A1J1IUJ7 A0A068TJS2 A0A026WII5 A0A034VPU7 A0A0A1XFV2 A0A1I8NSV1 A0A1B6GVV3 A0A336MGF4 T1PBD9 A0A232F5M0 K7IYA3 A0A0M9AAI4 A0A195C1R0 A0A1L8EEM7 A0A158NNF3 A0A1B6EYJ5 A0A1L8E2M4 P54385-2 A0A0P8ZYJ0 A0A0Q9WUT7 A0A0R1EBF8 A0A182QQS6 A0A1W4U8M3 A0A1J1IWE9 A0A034VKL6 A0A2M4CUS3 A0A0Q9WR91 A0A3B0KCM3 A0A1I8NSU8 I5ANX6 A0A2M4CT81 A0A0C9Q076 A0A0Q9X1Y5 A0A0A1X040 A0A1A9Y0I5 A0A1B0B2Z8 T1P987 A0A1A9W5Q9 D3TQR2 A0A1A9ZSU9 A0A0K8UFK5 A0A2M3ZZI4 A0A336KTS7 A0A182R8H6 A0A1L8EER7 F5HLL7 A0A2M3YYJ1 B3M317 A0A0L0BRA3 A0A182XF30 A0A0M3QYL6 B4N9E8 B4PL48 B3P8B7 P54385 A0A1W4UJV6 B4QSD4 U5EZ79 A0A3B0JQU1 B4HG10 Q298M7 B4G4C1 A0A182LW14 B4K740 B4M0C5 A0A182PG80 A0A023ETW6 A0A0L7RDB5 B4JRQ0 Q2KQ98 A0A2M3ZZC8 A0A182IW02 A0A182V234 A0A182UC31 A0A182HVJ8 A0A182YJQ2
A0A194RNK3 I4DJ42 A0A0K8TT23 E2C3C7 E9IPG3 A0A310SFZ1 A0A151X7U3 A0A2A3EPX9 A0A087ZS68 E2A6U2 A0A2J7PVJ1 A0A195FTW7 A0A154PIZ3 A0A158NNF9 W8BYK3 A0A1J1IUJ7 A0A068TJS2 A0A026WII5 A0A034VPU7 A0A0A1XFV2 A0A1I8NSV1 A0A1B6GVV3 A0A336MGF4 T1PBD9 A0A232F5M0 K7IYA3 A0A0M9AAI4 A0A195C1R0 A0A1L8EEM7 A0A158NNF3 A0A1B6EYJ5 A0A1L8E2M4 P54385-2 A0A0P8ZYJ0 A0A0Q9WUT7 A0A0R1EBF8 A0A182QQS6 A0A1W4U8M3 A0A1J1IWE9 A0A034VKL6 A0A2M4CUS3 A0A0Q9WR91 A0A3B0KCM3 A0A1I8NSU8 I5ANX6 A0A2M4CT81 A0A0C9Q076 A0A0Q9X1Y5 A0A0A1X040 A0A1A9Y0I5 A0A1B0B2Z8 T1P987 A0A1A9W5Q9 D3TQR2 A0A1A9ZSU9 A0A0K8UFK5 A0A2M3ZZI4 A0A336KTS7 A0A182R8H6 A0A1L8EER7 F5HLL7 A0A2M3YYJ1 B3M317 A0A0L0BRA3 A0A182XF30 A0A0M3QYL6 B4N9E8 B4PL48 B3P8B7 P54385 A0A1W4UJV6 B4QSD4 U5EZ79 A0A3B0JQU1 B4HG10 Q298M7 B4G4C1 A0A182LW14 B4K740 B4M0C5 A0A182PG80 A0A023ETW6 A0A0L7RDB5 B4JRQ0 Q2KQ98 A0A2M3ZZC8 A0A182IW02 A0A182V234 A0A182UC31 A0A182HVJ8 A0A182YJQ2
PDB
6DHN
E-value=0,
Score=2021
Ontologies
KEGG
PATHWAY
00220
Arginine biosynthesis - Bombyx mori (domestic silkworm)
00250 Alanine, aspartate and glutamate metabolism - Bombyx mori (domestic silkworm)
00471 D-Glutamine and D-glutamate metabolism - Bombyx mori (domestic silkworm)
00910 Nitrogen metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
00250 Alanine, aspartate and glutamate metabolism - Bombyx mori (domestic silkworm)
00471 D-Glutamine and D-glutamate metabolism - Bombyx mori (domestic silkworm)
00910 Nitrogen metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
GO
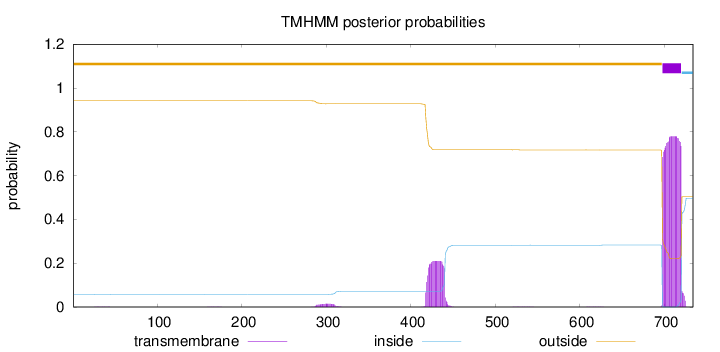
Topology
Subcellular location
Mitochondrion matrix
Length:
734
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.53334
Exp number, first 60 AAs:
0.00334
Total prob of N-in:
0.05795
outside
1 - 697
TMhelix
698 - 720
inside
721 - 734
Population Genetic Test Statistics
Pi
227.514607
Theta
178.991451
Tajima's D
0.810314
CLR
0.046095
CSRT
0.601369931503425
Interpretation
Possibly Positive selection
