Gene
KWMTBOMO03424 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006371
Annotation
Uncharacterized_protein_OBRU01_01696_[Operophtera_brumata]
Location in the cell
Extracellular Reliability : 1.722 Nuclear Reliability : 1.264
Sequence
CDS
ATGTTGCCACGGCCCCTCTCAGCGAGTACAATGCGGATTTTATTCAGTTTAGTGCTGGTAGCGTGCGCGGTCGACGCCGCAAAAAAGTTCGAGGGAACTTTGAGATCGGCAGCCGAGGCCCCACAAGAGGACAACTTAGCTATCGAATCTGCTTCTGCCGAACCGGCGGTTGTTGCTGCACCTCAAGACTATCAAAATCCTGGCGCTCCACCTCCTCCTAAAGATGAATTAGAATCTGAAGAACCCGCTGAAGAGCCTGCTGCTGTTCCCGAAACCGCATCCGAAGGAATACCACCGTCAGCTCCCAGTGGCGCTTCTGCACCATCGGCACCAGCTAATTCATTAGAAGAATGTGACCCCGAAATGATTGGCTTCGAATTAGTGACGGGGTACGTGTTCTCGGCGCCATCGCAAATTTTAGATGACATCCCCGGCACGCTGATGCTGACCGACTGCCTCGAGCAATGCCATGCTAACGACACCTGTCGCGCCGTAAATTACGAGACGGGACTTTGTGTGCTTTTCAGTTCTGATGCCGACCAACTGCCTGGTGCATTGACGAAATCGCAGTTCCCCGTATTCACTATATACGCTCAAAAATCTTGTTTGGGAGTTCGTCCGTGCGAACGCGCCTGGTGCTTCGATAGAGTGCGAGGCTACCACCTCAAGGGGCACGGCAAGAAAACACACACCGTTGGATCTCGGCAGATGTGCCTTGACCTTTGTCTCGGCGAAAACGAATTTGTTTGCAGATCTGCGAACTATAACAATAAAACCGGCGAGTGTGTGCTCTCTAACATGGACCGTATCACCTTGGCGGGAACCAACGCTTTTCAATCAAACGAAGACACCGACTATTTGGAGAACAACTGCGTTGAAGAGCCAACAAAATTGTGCGAGTTTAAAAAAATGGGTGGACGTATTCTTAAGACTGTTGATTCCGTATATCAAGACGTGCAGACTATTGAAGAATGCCGAGAACTTTGTCTAAATTCGCCTTTCCGTTGTCACTCATATGATCACGGTGATACTGGTGACCACGTCTGTCGTCTCTCTCATCACTCTCGGGCTACACTCGCTGATATCCAGGATCCTTATTTGGAAGTTCCCGAAGCAGCCACTTACGAACTGTCATCCTGTTACAACGTATCCATTGACTGTCGTGCTGGTGACATGGTTGCCAAAATTCAAACATCCAAATTGTTTGACGGCAAAATCTATGCTAAAGGAAGTCCAAACTCTTGCGTCGTAGATGTCAAACAGAGTCTTGAGTTTGAACTGCACATGGGTTATAACAATATCGAATGTAATGTTAAACAAAACGGACTTGGAAGGTATTTAAATGACGTTGTTATTCAACACCACGACACAATCGTAACTTCGTCTGATCTTGGTTTAGCGGTTACTTGTCAGTATGATTTGACAAACAAGACTGTTGCCAATGAAGTCGATCTTGGAATTCACGGAGACATTCAAACTGGTCTGTCCGAGGAAGTTATTGTAGACTCACCCAACGTAGCGATGAGAATCACAGACAGGAGTGGTGACGATACTATTGCTTCCGCTGAAGTTGGAGATCCATTGGCTCTCCGATTTGAAATTATGGATCCCAACTCACCATTCGAGATCTTCGTGCGAGAACTTGTTGCCATGGATGGTGTTGACTCCAGCGAAATCACGCTTATCGACAGCGATGGTTGCCCTACAGAACATTTTATCATGGGGCCACTATTCAAGTCCACTTTAAGCGGAAAGACGCTGCTATCACATTTCGACGCCTTCAAATTCCCTTCGTCAGAAGTTGTGCAATTCCGTGCCTTGGTGACTCCTTGTATGCCTACTTGCGAACCAGTGCAATGTGATGGCGGTCCGAATGAATTACGTTCAGTAATGTCATACGGTCGTAGGAAGAGGCGTTCTACATCTGCTACACCTACAGATGATATGCTTCTCGTGCAGACCATCCAGATTACTGATAAATTCGGATTCGACAAACAACGTGCTAAGAATGTCACAGAGGATGCAGTATATGTAAGAGAGACCGACGGAACTTGTGTGAATGCTTCAGGAGCCATGCTAGCTGCGGCCGCTTTCTTGGCAGCTCAACTAGTAGTACTTGCAGCGTGGACCTGCAGTTGGCAACGAAGACGTGCTGCTGCAAAAGCTGCCGAACTTCTTCCAGGCCCTAACGCACCCTCATCGCTGTGCAAGGTCTACGATGCGAGCTTCCCTCGTGCACATCAGAGGCACTTTTGA
Protein
MLPRPLSASTMRILFSLVLVACAVDAAKKFEGTLRSAAEAPQEDNLAIESASAEPAVVAAPQDYQNPGAPPPPKDELESEEPAEEPAAVPETASEGIPPSAPSGASAPSAPANSLEECDPEMIGFELVTGYVFSAPSQILDDIPGTLMLTDCLEQCHANDTCRAVNYETGLCVLFSSDADQLPGALTKSQFPVFTIYAQKSCLGVRPCERAWCFDRVRGYHLKGHGKKTHTVGSRQMCLDLCLGENEFVCRSANYNNKTGECVLSNMDRITLAGTNAFQSNEDTDYLENNCVEEPTKLCEFKKMGGRILKTVDSVYQDVQTIEECRELCLNSPFRCHSYDHGDTGDHVCRLSHHSRATLADIQDPYLEVPEAATYELSSCYNVSIDCRAGDMVAKIQTSKLFDGKIYAKGSPNSCVVDVKQSLEFELHMGYNNIECNVKQNGLGRYLNDVVIQHHDTIVTSSDLGLAVTCQYDLTNKTVANEVDLGIHGDIQTGLSEEVIVDSPNVAMRITDRSGDDTIASAEVGDPLALRFEIMDPNSPFEIFVRELVAMDGVDSSEITLIDSDGCPTEHFIMGPLFKSTLSGKTLLSHFDAFKFPSSEVVQFRALVTPCMPTCEPVQCDGGPNELRSVMSYGRRKRRSTSATPTDDMLLVQTIQITDKFGFDKQRAKNVTEDAVYVRETDGTCVNASGAMLAAAAFLAAQLVVLAAWTCSWQRRRAAAKAAELLPGPNAPSSLCKVYDASFPRAHQRHF
Summary
Uniprot
A0A2A4J5Y3
A0A2H1VQJ4
H9JA26
A0A0L7LTQ8
I4DIJ2
I4DM08
+ More
A0A194PXM5 A0A194RNJ8 A0A212F9C1 A0A1Y1KHG4 D6WEC2 A0A087ZXY2 Q171P7 A0A158P387 A0A195EZ81 A0A2C9H5R2 Q7QBU4 A0A0C9Q7Z9 A0A1I8NEV1 A0A084VR87 A0A1W4WMX9 A0A1I8QDM2 K7J2B0 J9KAI5 B4NA23 W8ATI6 B4LMX9 A0A2S2R5W8 A0A1J1HMP6 B4KT11 A0A0Q9X8J5 B4JSG6 A0A034VA94 A0A0K8V187 B4N9U9 B4PPJ2 A0A1W4UR70 B4HZF8 B3LWY2 B4R0T6 B3P5C3 Q9VAG2 A0A0M4EME8 B4LMY0 A0A3B0K9B5 A0A3B0KB54 B4GNR1 A0A0M4ESS7 A0A1B0ACN9 A0A1A9UM99 B4K8A0 A0A1A9XI83 B4KT12 B3M3A4 B4NIE3 B4LX02 Q9V9X0 B4QSN2 A0A1I8N6V2 B4IJ01 B4JG50 A0A3B0JG58 Q8IGS2 B3P534 B4JRW7 A0A1W4W708 B5DYT0 B4GYU9 A0A0K8TYA5 B4PNE6 A0A034VAC8 T1HKB1 A0A0A1X8W7 A0A0K8UC22 A0A1B0CQS2 A0A1B0AYB5 B4JRW8 A0A1A9W7X4 B4LX01 B4NIE6 B3M3A3 Q9V9X1 B3P535 B4QSN1 B5DYS9 B4PNE5 A0A1W4VW08 B4I033 A0A3B0JFL0 A0A3B0KC09 B4K8A2 A0A0L0BLS4 A0A1B0FF31 A0A0M4EKR6 A0A1W4WCW5 A0A034VBU4 A0A0P6HZU7 A0A164WWC9 A0A0P6EUA0
A0A194PXM5 A0A194RNJ8 A0A212F9C1 A0A1Y1KHG4 D6WEC2 A0A087ZXY2 Q171P7 A0A158P387 A0A195EZ81 A0A2C9H5R2 Q7QBU4 A0A0C9Q7Z9 A0A1I8NEV1 A0A084VR87 A0A1W4WMX9 A0A1I8QDM2 K7J2B0 J9KAI5 B4NA23 W8ATI6 B4LMX9 A0A2S2R5W8 A0A1J1HMP6 B4KT11 A0A0Q9X8J5 B4JSG6 A0A034VA94 A0A0K8V187 B4N9U9 B4PPJ2 A0A1W4UR70 B4HZF8 B3LWY2 B4R0T6 B3P5C3 Q9VAG2 A0A0M4EME8 B4LMY0 A0A3B0K9B5 A0A3B0KB54 B4GNR1 A0A0M4ESS7 A0A1B0ACN9 A0A1A9UM99 B4K8A0 A0A1A9XI83 B4KT12 B3M3A4 B4NIE3 B4LX02 Q9V9X0 B4QSN2 A0A1I8N6V2 B4IJ01 B4JG50 A0A3B0JG58 Q8IGS2 B3P534 B4JRW7 A0A1W4W708 B5DYT0 B4GYU9 A0A0K8TYA5 B4PNE6 A0A034VAC8 T1HKB1 A0A0A1X8W7 A0A0K8UC22 A0A1B0CQS2 A0A1B0AYB5 B4JRW8 A0A1A9W7X4 B4LX01 B4NIE6 B3M3A3 Q9V9X1 B3P535 B4QSN1 B5DYS9 B4PNE5 A0A1W4VW08 B4I033 A0A3B0JFL0 A0A3B0KC09 B4K8A2 A0A0L0BLS4 A0A1B0FF31 A0A0M4EKR6 A0A1W4WCW5 A0A034VBU4 A0A0P6HZU7 A0A164WWC9 A0A0P6EUA0
Pubmed
19121390
26227816
22651552
26354079
22118469
28004739
+ More
18362917 19820115 17510324 21347285 12364791 14747013 17210077 25315136 24438588 20075255 17994087 24495485 25348373 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 25830018 26108605
18362917 19820115 17510324 21347285 12364791 14747013 17210077 25315136 24438588 20075255 17994087 24495485 25348373 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 25830018 26108605
EMBL
NWSH01002817
PCG67485.1
ODYU01003825
SOQ43068.1
BABH01009281
JTDY01000100
+ More
KOB78868.1 AK401110 BAM17732.1 AK402326 BAM18948.1 KQ459589 KPI97509.1 KQ459896 KPJ19413.1 AGBW02009607 OWR50334.1 GEZM01083953 JAV60912.1 KQ971326 EFA00860.1 CH477449 EAT40707.1 ADTU01007829 ADTU01007830 ADTU01007831 ADTU01007832 ADTU01007833 ADTU01007834 ADTU01007835 ADTU01007836 ADTU01007837 ADTU01007838 KQ981905 KYN33615.1 AAAB01008859 EAA07511.5 GBYB01010278 GBYB01014249 JAG80045.1 JAG84016.1 ATLV01015531 ATLV01015532 ATLV01015533 ATLV01015534 KE525019 KFB40481.1 AAZX01001573 AAZX01005707 AAZX01006495 AAZX01013344 AAZX01017096 ABLF02031505 CH964232 EDW80666.1 GAMC01017048 JAB89507.1 CH940648 EDW62094.2 GGMS01016198 MBY85401.1 CVRI01000012 CRK89336.1 CH933808 EDW09531.2 KRG04726.1 CH916373 EDV94706.1 GAKP01019915 JAC39037.1 GDHF01019640 JAI32674.1 EDW80664.2 CM000160 EDW98242.1 CH480819 EDW53415.1 CH902617 EDV43821.1 CM000364 EDX14907.1 CH954182 EDV53173.1 AE014297 AY094725 AAF56948.1 AAM11078.1 AAS65226.1 CP012526 ALC46615.1 EDW62095.1 OUUW01000041 SPP89953.1 SPP89952.1 CH479186 EDW39387.1 ALC47847.1 CH933806 EDW16482.1 EDW09532.2 EDV43565.1 CH964272 EDW83725.1 CH940650 EDW67749.1 BT133074 AAF57159.2 AAN14274.2 AEX93159.1 EDX15129.1 CH480846 EDW50954.1 CH916369 EDV93617.1 OUUW01000005 SPP81125.1 BT001629 AAN71384.1 EDV52948.1 EDV94507.1 CM000070 EDY67640.2 CH479198 EDW27967.1 GDHF01032845 JAI19469.1 EDW99228.2 GAKP01019885 JAC39067.1 ACPB03011908 GBXI01007169 JAD07123.1 GDHF01028188 JAI24126.1 AJWK01023972 JXJN01005607 EDV94508.1 EDW67748.2 EDW83728.1 EDV43564.2 AAF57158.3 AHN57621.1 EDV52949.1 EDX15128.1 EDY67639.1 EDW99227.1 EDW53640.1 SPP81127.1 SPP81128.1 EDW16484.2 JRES01001677 KNC21006.1 CCAG010009783 ALC47846.1 GAKP01019919 GAKP01019916 JAC39033.1 GDIQ01011800 JAN82937.1 LRGB01001036 KZS13641.1 GDIQ01070787 JAN23950.1
KOB78868.1 AK401110 BAM17732.1 AK402326 BAM18948.1 KQ459589 KPI97509.1 KQ459896 KPJ19413.1 AGBW02009607 OWR50334.1 GEZM01083953 JAV60912.1 KQ971326 EFA00860.1 CH477449 EAT40707.1 ADTU01007829 ADTU01007830 ADTU01007831 ADTU01007832 ADTU01007833 ADTU01007834 ADTU01007835 ADTU01007836 ADTU01007837 ADTU01007838 KQ981905 KYN33615.1 AAAB01008859 EAA07511.5 GBYB01010278 GBYB01014249 JAG80045.1 JAG84016.1 ATLV01015531 ATLV01015532 ATLV01015533 ATLV01015534 KE525019 KFB40481.1 AAZX01001573 AAZX01005707 AAZX01006495 AAZX01013344 AAZX01017096 ABLF02031505 CH964232 EDW80666.1 GAMC01017048 JAB89507.1 CH940648 EDW62094.2 GGMS01016198 MBY85401.1 CVRI01000012 CRK89336.1 CH933808 EDW09531.2 KRG04726.1 CH916373 EDV94706.1 GAKP01019915 JAC39037.1 GDHF01019640 JAI32674.1 EDW80664.2 CM000160 EDW98242.1 CH480819 EDW53415.1 CH902617 EDV43821.1 CM000364 EDX14907.1 CH954182 EDV53173.1 AE014297 AY094725 AAF56948.1 AAM11078.1 AAS65226.1 CP012526 ALC46615.1 EDW62095.1 OUUW01000041 SPP89953.1 SPP89952.1 CH479186 EDW39387.1 ALC47847.1 CH933806 EDW16482.1 EDW09532.2 EDV43565.1 CH964272 EDW83725.1 CH940650 EDW67749.1 BT133074 AAF57159.2 AAN14274.2 AEX93159.1 EDX15129.1 CH480846 EDW50954.1 CH916369 EDV93617.1 OUUW01000005 SPP81125.1 BT001629 AAN71384.1 EDV52948.1 EDV94507.1 CM000070 EDY67640.2 CH479198 EDW27967.1 GDHF01032845 JAI19469.1 EDW99228.2 GAKP01019885 JAC39067.1 ACPB03011908 GBXI01007169 JAD07123.1 GDHF01028188 JAI24126.1 AJWK01023972 JXJN01005607 EDV94508.1 EDW67748.2 EDW83728.1 EDV43564.2 AAF57158.3 AHN57621.1 EDV52949.1 EDX15128.1 EDY67639.1 EDW99227.1 EDW53640.1 SPP81127.1 SPP81128.1 EDW16484.2 JRES01001677 KNC21006.1 CCAG010009783 ALC47846.1 GAKP01019919 GAKP01019916 JAC39033.1 GDIQ01011800 JAN82937.1 LRGB01001036 KZS13641.1 GDIQ01070787 JAN23950.1
Proteomes
UP000218220
UP000005204
UP000037510
UP000053268
UP000053240
UP000007151
+ More
UP000007266 UP000005203 UP000008820 UP000005205 UP000078541 UP000075903 UP000007062 UP000095301 UP000030765 UP000192223 UP000095300 UP000002358 UP000007819 UP000007798 UP000008792 UP000183832 UP000009192 UP000001070 UP000002282 UP000192221 UP000001292 UP000007801 UP000000304 UP000008711 UP000000803 UP000092553 UP000268350 UP000008744 UP000092445 UP000078200 UP000092443 UP000001819 UP000015103 UP000092461 UP000092460 UP000091820 UP000037069 UP000092444 UP000076858
UP000007266 UP000005203 UP000008820 UP000005205 UP000078541 UP000075903 UP000007062 UP000095301 UP000030765 UP000192223 UP000095300 UP000002358 UP000007819 UP000007798 UP000008792 UP000183832 UP000009192 UP000001070 UP000002282 UP000192221 UP000001292 UP000007801 UP000000304 UP000008711 UP000000803 UP000092553 UP000268350 UP000008744 UP000092445 UP000078200 UP000092443 UP000001819 UP000015103 UP000092461 UP000092460 UP000091820 UP000037069 UP000092444 UP000076858
ProteinModelPortal
A0A2A4J5Y3
A0A2H1VQJ4
H9JA26
A0A0L7LTQ8
I4DIJ2
I4DM08
+ More
A0A194PXM5 A0A194RNJ8 A0A212F9C1 A0A1Y1KHG4 D6WEC2 A0A087ZXY2 Q171P7 A0A158P387 A0A195EZ81 A0A2C9H5R2 Q7QBU4 A0A0C9Q7Z9 A0A1I8NEV1 A0A084VR87 A0A1W4WMX9 A0A1I8QDM2 K7J2B0 J9KAI5 B4NA23 W8ATI6 B4LMX9 A0A2S2R5W8 A0A1J1HMP6 B4KT11 A0A0Q9X8J5 B4JSG6 A0A034VA94 A0A0K8V187 B4N9U9 B4PPJ2 A0A1W4UR70 B4HZF8 B3LWY2 B4R0T6 B3P5C3 Q9VAG2 A0A0M4EME8 B4LMY0 A0A3B0K9B5 A0A3B0KB54 B4GNR1 A0A0M4ESS7 A0A1B0ACN9 A0A1A9UM99 B4K8A0 A0A1A9XI83 B4KT12 B3M3A4 B4NIE3 B4LX02 Q9V9X0 B4QSN2 A0A1I8N6V2 B4IJ01 B4JG50 A0A3B0JG58 Q8IGS2 B3P534 B4JRW7 A0A1W4W708 B5DYT0 B4GYU9 A0A0K8TYA5 B4PNE6 A0A034VAC8 T1HKB1 A0A0A1X8W7 A0A0K8UC22 A0A1B0CQS2 A0A1B0AYB5 B4JRW8 A0A1A9W7X4 B4LX01 B4NIE6 B3M3A3 Q9V9X1 B3P535 B4QSN1 B5DYS9 B4PNE5 A0A1W4VW08 B4I033 A0A3B0JFL0 A0A3B0KC09 B4K8A2 A0A0L0BLS4 A0A1B0FF31 A0A0M4EKR6 A0A1W4WCW5 A0A034VBU4 A0A0P6HZU7 A0A164WWC9 A0A0P6EUA0
A0A194PXM5 A0A194RNJ8 A0A212F9C1 A0A1Y1KHG4 D6WEC2 A0A087ZXY2 Q171P7 A0A158P387 A0A195EZ81 A0A2C9H5R2 Q7QBU4 A0A0C9Q7Z9 A0A1I8NEV1 A0A084VR87 A0A1W4WMX9 A0A1I8QDM2 K7J2B0 J9KAI5 B4NA23 W8ATI6 B4LMX9 A0A2S2R5W8 A0A1J1HMP6 B4KT11 A0A0Q9X8J5 B4JSG6 A0A034VA94 A0A0K8V187 B4N9U9 B4PPJ2 A0A1W4UR70 B4HZF8 B3LWY2 B4R0T6 B3P5C3 Q9VAG2 A0A0M4EME8 B4LMY0 A0A3B0K9B5 A0A3B0KB54 B4GNR1 A0A0M4ESS7 A0A1B0ACN9 A0A1A9UM99 B4K8A0 A0A1A9XI83 B4KT12 B3M3A4 B4NIE3 B4LX02 Q9V9X0 B4QSN2 A0A1I8N6V2 B4IJ01 B4JG50 A0A3B0JG58 Q8IGS2 B3P534 B4JRW7 A0A1W4W708 B5DYT0 B4GYU9 A0A0K8TYA5 B4PNE6 A0A034VAC8 T1HKB1 A0A0A1X8W7 A0A0K8UC22 A0A1B0CQS2 A0A1B0AYB5 B4JRW8 A0A1A9W7X4 B4LX01 B4NIE6 B3M3A3 Q9V9X1 B3P535 B4QSN1 B5DYS9 B4PNE5 A0A1W4VW08 B4I033 A0A3B0JFL0 A0A3B0KC09 B4K8A2 A0A0L0BLS4 A0A1B0FF31 A0A0M4EKR6 A0A1W4WCW5 A0A034VBU4 A0A0P6HZU7 A0A164WWC9 A0A0P6EUA0
Ontologies
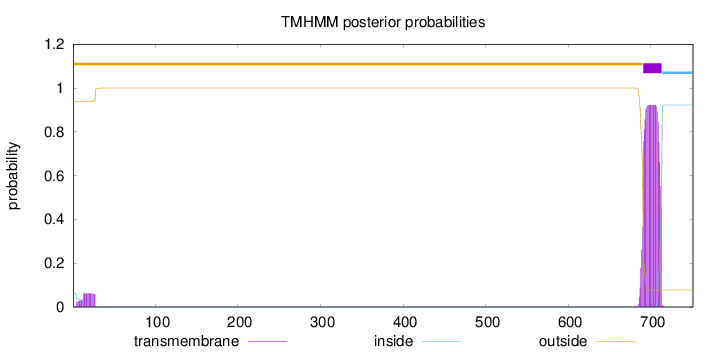
Topology
SignalP
Position: 1 - 26,
Likelihood: 0.957163
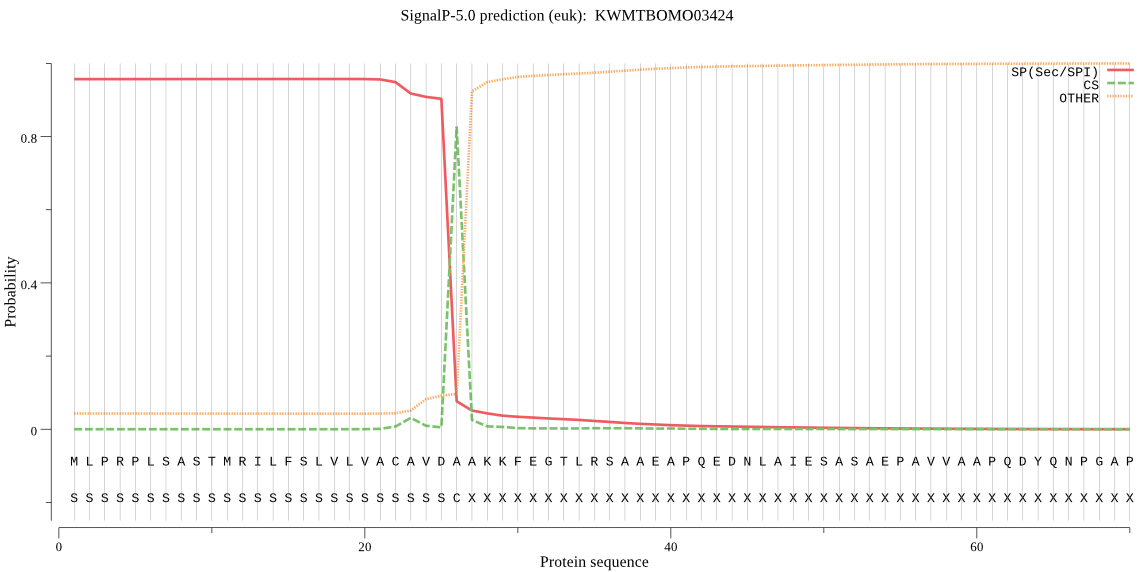
Length:
751
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.57657
Exp number, first 60 AAs:
1.15751
Total prob of N-in:
0.06205
outside
1 - 690
TMhelix
691 - 713
inside
714 - 751
Population Genetic Test Statistics
Pi
201.860551
Theta
194.305977
Tajima's D
0.224708
CLR
0.585843
CSRT
0.43892805359732
Interpretation
Uncertain
