Pre Gene Modal
BGIBMGA014287
Annotation
PREDICTED:_acetyl-coenzyme_A_synthetase_1-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.385
Sequence
CDS
ATGTTGGAACATATCCGACAGTGGAATGACGAAATTGCAATAGAAAATGCTGATACAGGTGAAAAACTATCGTACAAAGAAATCGCACAGTATGTTGTGAACCTTTCCGCGTCTTTGACGAGCCTTGGAGTCGGAAGAGGAGATACAGTGGCACTCGGGTCGCAAAAACACAATGAATTTGTTCCGACTTTGCTTGCAATTGTATTGACTGGAGCCACCTATACACCATAA
Protein
MLEHIRQWNDEIAIENADTGEKLSYKEIAQYVVNLSASLTSLGVGRGDTVALGSQKHNEFVPTLLAIVLTGATYTP
Summary
Similarity
Belongs to the ATP-dependent AMP-binding enzyme family.
Belongs to the peptidase S1 family.
Belongs to the peptidase S1 family.
Uniprot
H9JXM0
A0A2A4J3S6
A0A2H1V0T1
A0A0L7LAG5
A0A0L7LP26
A0A0L7L6L8
+ More
A0A0L7LH10 A0A0L7LG20 A0A194QBD3 A0A0L7L784 A0A0L7L765 A0A2H1WJN9 A0A2H1V0C4 A0A2H1VKC8 G8GE17 H9J2F6 A0A194PTD5 A0A212FIH9 A0A2A4JMM7 A0A194RLN8 A0A194PGU6 A0A194QZM0 A0A0L7LHE0 A0A212FII9 A0A2H1WNI5 A0A194PZH5 A0A194QZV5 A0A2A4JNM0 A0A194R0D8 A0A194QRL9 I4DPZ5 A0A2G7EQP5 A0A0L7KQP2 A0A260Q0T6 A0A194PUC3 A0A2A4J232 A0A285BPA9 A0A194R547 A0A3G2IPV5 A0A3N4ZH13 A0A0B5EQR2 A0A2A9G3C5 A0A2H1VTP5 A0A352Q3R9 A0A2A4JYH7 A0A194PUZ5 A0A0M8XQK3 A0A1H3QLS3 A0A2N1WWR4 A0A1G6UP45 A0A2W1BTP3 A0A2M9FG45 A0A2H1WXP8 A0A0F5VWY7 A0A2N7W0X2 A0A327U5M0 A0A2Z5J6H9 A0A255PP22 A0A0U5LKR0 A0A0L8LP41 A0A1Q4TJV9 A0A1J0R3L9 A0A3E0HEX3 A0A1C6U9D9 M2QKS4 A0A1C6RC28 A0A212ETT6 A0A150N3K0 A0A0K1J6S1 A0A370F929 A0A2H1VNT1 A0A1J1EC51 A0A194PT23 A0A2N3Y0L3 A0A0T1W5J2 A0A209BMT4 A0A193C668 A0A0K6IAU5 A0A1A1WZF6 A0A3M1W2W2 A0A1C5BRG6 A0A1V2QNC9 A0A1C6SCY6 M3A0X4 A0A3M2LET6 A0A1D3DNR8 A0A2X0K1N1 A0A1S1NE22 A0A081XYT4 A0A2K8GMH6 Q8KLL5 A0A1V9BD87 A0A3E0GZG9 A0A0W0LMV6 A0A1Q4VSJ2 A0A094RW75 A0A3M4FG74 I4DPZ6 A0A2N3K8A8
A0A0L7LH10 A0A0L7LG20 A0A194QBD3 A0A0L7L784 A0A0L7L765 A0A2H1WJN9 A0A2H1V0C4 A0A2H1VKC8 G8GE17 H9J2F6 A0A194PTD5 A0A212FIH9 A0A2A4JMM7 A0A194RLN8 A0A194PGU6 A0A194QZM0 A0A0L7LHE0 A0A212FII9 A0A2H1WNI5 A0A194PZH5 A0A194QZV5 A0A2A4JNM0 A0A194R0D8 A0A194QRL9 I4DPZ5 A0A2G7EQP5 A0A0L7KQP2 A0A260Q0T6 A0A194PUC3 A0A2A4J232 A0A285BPA9 A0A194R547 A0A3G2IPV5 A0A3N4ZH13 A0A0B5EQR2 A0A2A9G3C5 A0A2H1VTP5 A0A352Q3R9 A0A2A4JYH7 A0A194PUZ5 A0A0M8XQK3 A0A1H3QLS3 A0A2N1WWR4 A0A1G6UP45 A0A2W1BTP3 A0A2M9FG45 A0A2H1WXP8 A0A0F5VWY7 A0A2N7W0X2 A0A327U5M0 A0A2Z5J6H9 A0A255PP22 A0A0U5LKR0 A0A0L8LP41 A0A1Q4TJV9 A0A1J0R3L9 A0A3E0HEX3 A0A1C6U9D9 M2QKS4 A0A1C6RC28 A0A212ETT6 A0A150N3K0 A0A0K1J6S1 A0A370F929 A0A2H1VNT1 A0A1J1EC51 A0A194PT23 A0A2N3Y0L3 A0A0T1W5J2 A0A209BMT4 A0A193C668 A0A0K6IAU5 A0A1A1WZF6 A0A3M1W2W2 A0A1C5BRG6 A0A1V2QNC9 A0A1C6SCY6 M3A0X4 A0A3M2LET6 A0A1D3DNR8 A0A2X0K1N1 A0A1S1NE22 A0A081XYT4 A0A2K8GMH6 Q8KLL5 A0A1V9BD87 A0A3E0GZG9 A0A0W0LMV6 A0A1Q4VSJ2 A0A094RW75 A0A3M4FG74 I4DPZ6 A0A2N3K8A8
Pubmed
EMBL
BABH01044598
NWSH01003607
PCG66062.1
ODYU01000123
SOQ34366.1
JTDY01001941
+ More
KOB72488.1 JTDY01000482 KOB76976.1 JTDY01002678 KOB70956.1 JTDY01001219 KOB74496.1 KOB74498.1 KQ459249 KPJ02300.1 JTDY01002474 KOB71343.1 KOB71342.1 ODYU01009054 SOQ53186.1 ODYU01000100 SOQ34295.1 ODYU01002875 SOQ40902.1 JN127350 AET05796.1 BABH01007891 KQ459593 KPI96567.1 AGBW02008380 OWR53543.1 NWSH01001030 PCG73036.1 KQ460045 KPJ18230.1 KQ459604 KPI92263.1 KQ460890 KPJ10988.1 JTDY01001065 KOB74983.1 OWR53544.1 ODYU01009511 SOQ54004.1 KPI96565.1 KPJ10987.1 PCG73033.1 KPJ10989.1 KQ461181 KPJ07625.1 AK403860 BAM19985.1 PEFT01000001 PIG70670.1 JTDY01007153 KOB65430.1 NOZM01000004 OZE68710.1 KPI96573.1 NWSH01003686 PCG65919.1 OAOJ01000001 SNX56806.1 KPJ10986.1 RKRB01000001 RPF31434.1 CP010519 AJE80497.1 PDJK01000001 PFG57310.1 ODYU01004352 SOQ44136.1 DNIW01000129 HBD11853.1 NWSH01000380 PCG76826.1 KPI96564.1 LGEC01000105 KOX11032.1 FNOK01000048 SDZ14317.1 PGYX01000001 PKM01163.1 FMYS01000006 SDD42325.1 KZ149903 PZC78438.1 PEBB01000071 PJK22428.1 ODYU01011856 SOQ57839.1 JXTE01000029 KKD06322.1 PNYA01000002 PMS23068.1 QLLV01000017 RAJ50021.1 CP027306 AXE75899.1 NPKF01000001 OYP17903.1 LN997842 CUW28146.1 LGUS01000061 LMWZ01000003 KOG39887.1 KUO01030.1 JMHS01000024 OKH73598.1 KX708431 APD72103.1 QUNO01000009 REH43723.1 FMIC01000002 SCL50715.1 ANMG01000033 EMD26452.1 FMHT01000003 SCL14711.1 AGBW02012531 OWR44889.1 LQYX01000001 KYD31244.1 CP011072 AKU12394.1 QQAT01000016 RDI20464.1 ODYU01003402 SOQ42112.1 AP014940 BAV97564.1 KPI96566.1 PJNB01000001 PKW16455.1 LMEZ01000008 KQY03935.1 NEVE01000034 OWA06208.1 CP016174 ANN20076.1 CYHF01000014 CUB00447.1 LZHW01000101 OBF11851.1 RFHH01000039 RMG48180.1 FMDK01000013 SCF61112.1 MTQP01000055 ONI89757.1 FMHW01000002 SCL27341.1 AOEX01000025 EME66209.1 RFFH01000001 RMI35083.1 ASHX02000001 OEJ93967.1 QKYN01000093 RAG83175.1 MLQM01000148 OHU98270.1 JFCB01000001 KES08707.1 KY654519 ARU08075.1 U82965 AAM80537.1 NADR01000003 OQP07350.1 QUNO01000018 REH35336.1 LKGY01000123 KTC11023.1 LWKZ01000024 OKI00759.1 JQOF01000026 KGA39752.1 RBQQ01000043 RMP67011.1 AK403861 BAM19986.1 PITL01000002 PKR46790.1
KOB72488.1 JTDY01000482 KOB76976.1 JTDY01002678 KOB70956.1 JTDY01001219 KOB74496.1 KOB74498.1 KQ459249 KPJ02300.1 JTDY01002474 KOB71343.1 KOB71342.1 ODYU01009054 SOQ53186.1 ODYU01000100 SOQ34295.1 ODYU01002875 SOQ40902.1 JN127350 AET05796.1 BABH01007891 KQ459593 KPI96567.1 AGBW02008380 OWR53543.1 NWSH01001030 PCG73036.1 KQ460045 KPJ18230.1 KQ459604 KPI92263.1 KQ460890 KPJ10988.1 JTDY01001065 KOB74983.1 OWR53544.1 ODYU01009511 SOQ54004.1 KPI96565.1 KPJ10987.1 PCG73033.1 KPJ10989.1 KQ461181 KPJ07625.1 AK403860 BAM19985.1 PEFT01000001 PIG70670.1 JTDY01007153 KOB65430.1 NOZM01000004 OZE68710.1 KPI96573.1 NWSH01003686 PCG65919.1 OAOJ01000001 SNX56806.1 KPJ10986.1 RKRB01000001 RPF31434.1 CP010519 AJE80497.1 PDJK01000001 PFG57310.1 ODYU01004352 SOQ44136.1 DNIW01000129 HBD11853.1 NWSH01000380 PCG76826.1 KPI96564.1 LGEC01000105 KOX11032.1 FNOK01000048 SDZ14317.1 PGYX01000001 PKM01163.1 FMYS01000006 SDD42325.1 KZ149903 PZC78438.1 PEBB01000071 PJK22428.1 ODYU01011856 SOQ57839.1 JXTE01000029 KKD06322.1 PNYA01000002 PMS23068.1 QLLV01000017 RAJ50021.1 CP027306 AXE75899.1 NPKF01000001 OYP17903.1 LN997842 CUW28146.1 LGUS01000061 LMWZ01000003 KOG39887.1 KUO01030.1 JMHS01000024 OKH73598.1 KX708431 APD72103.1 QUNO01000009 REH43723.1 FMIC01000002 SCL50715.1 ANMG01000033 EMD26452.1 FMHT01000003 SCL14711.1 AGBW02012531 OWR44889.1 LQYX01000001 KYD31244.1 CP011072 AKU12394.1 QQAT01000016 RDI20464.1 ODYU01003402 SOQ42112.1 AP014940 BAV97564.1 KPI96566.1 PJNB01000001 PKW16455.1 LMEZ01000008 KQY03935.1 NEVE01000034 OWA06208.1 CP016174 ANN20076.1 CYHF01000014 CUB00447.1 LZHW01000101 OBF11851.1 RFHH01000039 RMG48180.1 FMDK01000013 SCF61112.1 MTQP01000055 ONI89757.1 FMHW01000002 SCL27341.1 AOEX01000025 EME66209.1 RFFH01000001 RMI35083.1 ASHX02000001 OEJ93967.1 QKYN01000093 RAG83175.1 MLQM01000148 OHU98270.1 JFCB01000001 KES08707.1 KY654519 ARU08075.1 U82965 AAM80537.1 NADR01000003 OQP07350.1 QUNO01000018 REH35336.1 LKGY01000123 KTC11023.1 LWKZ01000024 OKI00759.1 JQOF01000026 KGA39752.1 RBQQ01000043 RMP67011.1 AK403861 BAM19986.1 PITL01000002 PKR46790.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053268
UP000007151
UP000053240
+ More
UP000229013 UP000216557 UP000219027 UP000276963 UP000031523 UP000243542 UP000264566 UP000037694 UP000199529 UP000198991 UP000230216 UP000033641 UP000235616 UP000249176 UP000252698 UP000216770 UP000062854 UP000037251 UP000052992 UP000186656 UP000256269 UP000199343 UP000014137 UP000199699 UP000075654 UP000066621 UP000255116 UP000218824 UP000233786 UP000051127 UP000195919 UP000093695 UP000183649 UP000092314 UP000268325 UP000198886 UP000188617 UP000198959 UP000011731 UP000279275 UP000095329 UP000248889 UP000179734 UP000028341 UP000192323 UP000055023 UP000185893 UP000029447 UP000267110 UP000233474
UP000229013 UP000216557 UP000219027 UP000276963 UP000031523 UP000243542 UP000264566 UP000037694 UP000199529 UP000198991 UP000230216 UP000033641 UP000235616 UP000249176 UP000252698 UP000216770 UP000062854 UP000037251 UP000052992 UP000186656 UP000256269 UP000199343 UP000014137 UP000199699 UP000075654 UP000066621 UP000255116 UP000218824 UP000233786 UP000051127 UP000195919 UP000093695 UP000183649 UP000092314 UP000268325 UP000198886 UP000188617 UP000198959 UP000011731 UP000279275 UP000095329 UP000248889 UP000179734 UP000028341 UP000192323 UP000055023 UP000185893 UP000029447 UP000267110 UP000233474
Pfam
PF13193 AMP-binding_C
+ More
PF00501 AMP-binding
PF00550 PP-binding
PF00975 Thioesterase
PF00668 Condensation
PF00296 Bac_luciferase
PF08241 Methyltransf_11
PF00089 Trypsin
PF08242 Methyltransf_12
PF04299 FMN_bind_2
PF07993 NAD_binding_4
PF02801 Ketoacyl-synt_C
PF14765 PS-DH
PF13649 Methyltransf_25
PF08659 KR
PF00109 ketoacyl-synt
PF00698 Acyl_transf_1
PF16197 KAsynt_C_assoc
PF00501 AMP-binding
PF00550 PP-binding
PF00975 Thioesterase
PF00668 Condensation
PF00296 Bac_luciferase
PF08241 Methyltransf_11
PF00089 Trypsin
PF08242 Methyltransf_12
PF04299 FMN_bind_2
PF07993 NAD_binding_4
PF02801 Ketoacyl-synt_C
PF14765 PS-DH
PF13649 Methyltransf_25
PF08659 KR
PF00109 ketoacyl-synt
PF00698 Acyl_transf_1
PF16197 KAsynt_C_assoc
Interpro
IPR020845
AMP-binding_CS
+ More
IPR042099 AMP-dep_Synthh-like_sf
IPR000873 AMP-dep_Synth/Lig
IPR025110 AMP-bd_C
IPR036691 Endo/exonu/phosph_ase_sf
IPR010071 AA_adenyl_domain
IPR020459 AMP-binding
IPR036736 ACP-like_sf
IPR009081 PP-bd_ACP
IPR020806 PKS_PP-bd
IPR020802 PKS_thioesterase
IPR023213 CAT-like_dom_sf
IPR006162 Ppantetheine_attach_site
IPR001031 Thioesterase
IPR001242 Condensatn
IPR029058 AB_hydrolase
IPR029063 SAM-dependent_MTases
IPR024011 Biosynth_lucif-like_mOase_dom
IPR013216 Methyltransf_11
IPR036661 Luciferase-like_sf
IPR011251 Luciferase-like_dom
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR033116 TRYPSIN_SER
IPR013217 Methyltransf_12
IPR012349 Split_barrel_FMN-bd
IPR007396 TR_PAI2-type
IPR010060 NRPS_synth
IPR013120 Male_sterile_NAD-bd
IPR010080 Thioester_reductase-like_dom
IPR036291 NAD(P)-bd_dom_sf
IPR006342 FkbM_mtfrase
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR020807 PKS_dehydratase
IPR001227 Ac_transferase_dom_sf
IPR013968 PKS_KR
IPR020843 PKS_ER
IPR014030 Ketoacyl_synth_N
IPR016039 Thiolase-like
IPR020801 PKS_acyl_transferase
IPR002364 Quin_OxRdtase/zeta-crystal_CS
IPR042104 PKS_dehydratase_sf
IPR011032 GroES-like_sf
IPR032821 KAsynt_C_assoc
IPR014043 Acyl_transferase
IPR016036 Malonyl_transacylase_ACP-bd
IPR041698 Methyltransf_25
IPR016035 Acyl_Trfase/lysoPLipase
IPR014031 Ketoacyl_synth_C
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR017529 AcylCoA_ligase_PEP_1
IPR042099 AMP-dep_Synthh-like_sf
IPR000873 AMP-dep_Synth/Lig
IPR025110 AMP-bd_C
IPR036691 Endo/exonu/phosph_ase_sf
IPR010071 AA_adenyl_domain
IPR020459 AMP-binding
IPR036736 ACP-like_sf
IPR009081 PP-bd_ACP
IPR020806 PKS_PP-bd
IPR020802 PKS_thioesterase
IPR023213 CAT-like_dom_sf
IPR006162 Ppantetheine_attach_site
IPR001031 Thioesterase
IPR001242 Condensatn
IPR029058 AB_hydrolase
IPR029063 SAM-dependent_MTases
IPR024011 Biosynth_lucif-like_mOase_dom
IPR013216 Methyltransf_11
IPR036661 Luciferase-like_sf
IPR011251 Luciferase-like_dom
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR033116 TRYPSIN_SER
IPR013217 Methyltransf_12
IPR012349 Split_barrel_FMN-bd
IPR007396 TR_PAI2-type
IPR010060 NRPS_synth
IPR013120 Male_sterile_NAD-bd
IPR010080 Thioester_reductase-like_dom
IPR036291 NAD(P)-bd_dom_sf
IPR006342 FkbM_mtfrase
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR020807 PKS_dehydratase
IPR001227 Ac_transferase_dom_sf
IPR013968 PKS_KR
IPR020843 PKS_ER
IPR014030 Ketoacyl_synth_N
IPR016039 Thiolase-like
IPR020801 PKS_acyl_transferase
IPR002364 Quin_OxRdtase/zeta-crystal_CS
IPR042104 PKS_dehydratase_sf
IPR011032 GroES-like_sf
IPR032821 KAsynt_C_assoc
IPR014043 Acyl_transferase
IPR016036 Malonyl_transacylase_ACP-bd
IPR041698 Methyltransf_25
IPR016035 Acyl_Trfase/lysoPLipase
IPR014031 Ketoacyl_synth_C
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR017529 AcylCoA_ligase_PEP_1
SUPFAM
Gene 3D
ProteinModelPortal
H9JXM0
A0A2A4J3S6
A0A2H1V0T1
A0A0L7LAG5
A0A0L7LP26
A0A0L7L6L8
+ More
A0A0L7LH10 A0A0L7LG20 A0A194QBD3 A0A0L7L784 A0A0L7L765 A0A2H1WJN9 A0A2H1V0C4 A0A2H1VKC8 G8GE17 H9J2F6 A0A194PTD5 A0A212FIH9 A0A2A4JMM7 A0A194RLN8 A0A194PGU6 A0A194QZM0 A0A0L7LHE0 A0A212FII9 A0A2H1WNI5 A0A194PZH5 A0A194QZV5 A0A2A4JNM0 A0A194R0D8 A0A194QRL9 I4DPZ5 A0A2G7EQP5 A0A0L7KQP2 A0A260Q0T6 A0A194PUC3 A0A2A4J232 A0A285BPA9 A0A194R547 A0A3G2IPV5 A0A3N4ZH13 A0A0B5EQR2 A0A2A9G3C5 A0A2H1VTP5 A0A352Q3R9 A0A2A4JYH7 A0A194PUZ5 A0A0M8XQK3 A0A1H3QLS3 A0A2N1WWR4 A0A1G6UP45 A0A2W1BTP3 A0A2M9FG45 A0A2H1WXP8 A0A0F5VWY7 A0A2N7W0X2 A0A327U5M0 A0A2Z5J6H9 A0A255PP22 A0A0U5LKR0 A0A0L8LP41 A0A1Q4TJV9 A0A1J0R3L9 A0A3E0HEX3 A0A1C6U9D9 M2QKS4 A0A1C6RC28 A0A212ETT6 A0A150N3K0 A0A0K1J6S1 A0A370F929 A0A2H1VNT1 A0A1J1EC51 A0A194PT23 A0A2N3Y0L3 A0A0T1W5J2 A0A209BMT4 A0A193C668 A0A0K6IAU5 A0A1A1WZF6 A0A3M1W2W2 A0A1C5BRG6 A0A1V2QNC9 A0A1C6SCY6 M3A0X4 A0A3M2LET6 A0A1D3DNR8 A0A2X0K1N1 A0A1S1NE22 A0A081XYT4 A0A2K8GMH6 Q8KLL5 A0A1V9BD87 A0A3E0GZG9 A0A0W0LMV6 A0A1Q4VSJ2 A0A094RW75 A0A3M4FG74 I4DPZ6 A0A2N3K8A8
A0A0L7LH10 A0A0L7LG20 A0A194QBD3 A0A0L7L784 A0A0L7L765 A0A2H1WJN9 A0A2H1V0C4 A0A2H1VKC8 G8GE17 H9J2F6 A0A194PTD5 A0A212FIH9 A0A2A4JMM7 A0A194RLN8 A0A194PGU6 A0A194QZM0 A0A0L7LHE0 A0A212FII9 A0A2H1WNI5 A0A194PZH5 A0A194QZV5 A0A2A4JNM0 A0A194R0D8 A0A194QRL9 I4DPZ5 A0A2G7EQP5 A0A0L7KQP2 A0A260Q0T6 A0A194PUC3 A0A2A4J232 A0A285BPA9 A0A194R547 A0A3G2IPV5 A0A3N4ZH13 A0A0B5EQR2 A0A2A9G3C5 A0A2H1VTP5 A0A352Q3R9 A0A2A4JYH7 A0A194PUZ5 A0A0M8XQK3 A0A1H3QLS3 A0A2N1WWR4 A0A1G6UP45 A0A2W1BTP3 A0A2M9FG45 A0A2H1WXP8 A0A0F5VWY7 A0A2N7W0X2 A0A327U5M0 A0A2Z5J6H9 A0A255PP22 A0A0U5LKR0 A0A0L8LP41 A0A1Q4TJV9 A0A1J0R3L9 A0A3E0HEX3 A0A1C6U9D9 M2QKS4 A0A1C6RC28 A0A212ETT6 A0A150N3K0 A0A0K1J6S1 A0A370F929 A0A2H1VNT1 A0A1J1EC51 A0A194PT23 A0A2N3Y0L3 A0A0T1W5J2 A0A209BMT4 A0A193C668 A0A0K6IAU5 A0A1A1WZF6 A0A3M1W2W2 A0A1C5BRG6 A0A1V2QNC9 A0A1C6SCY6 M3A0X4 A0A3M2LET6 A0A1D3DNR8 A0A2X0K1N1 A0A1S1NE22 A0A081XYT4 A0A2K8GMH6 Q8KLL5 A0A1V9BD87 A0A3E0GZG9 A0A0W0LMV6 A0A1Q4VSJ2 A0A094RW75 A0A3M4FG74 I4DPZ6 A0A2N3K8A8
PDB
6P1J
E-value=0.000217372,
Score=99
Ontologies
GO
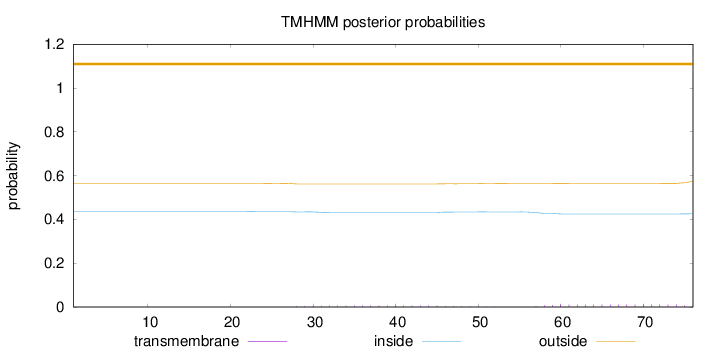
Topology
Length:
76
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.32096
Exp number, first 60 AAs:
0.15959
Total prob of N-in:
0.43537
outside
1 - 76
Population Genetic Test Statistics
Pi
256.426974
Theta
167.47086
Tajima's D
1.67068
CLR
0
CSRT
0.819609019549023
Interpretation
Uncertain
