Gene
KWMTBOMO03418
Pre Gene Modal
BGIBMGA006503
Annotation
PREDICTED:_lachesin-like_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.64 PlasmaMembrane Reliability : 1.684
Sequence
CDS
ATGAGCAAATACGGTCGTGAGTTACCTAGTGGGCAGAATGTTGTCTCAGTAGCACCTATAGAAGTTCCAGACGAATCAATGTACCCAAGATTCGCGGAGCCCATACCGAATGTCACGGTCACTATTGGCCGCGACGCTCTGTTGGCGTGCGTCGTTGAGAACCTGAGAGGTTTTAAGGTGGCATGGGTGCGAATGGACACCCAAACTATTCTAAGTATTCATCACAATATAATAACGCAGAATTCTCGCATCAGCCTGTCTTATAACGACCACCGTTCGTGGTACCTGCACATTAAAAACGTTCAGGAGGTCGATCGCGGTTGGTACATGTGCCAGGTTAACACGGACCCGATGCATTTTAGAAAAGGCTACTTGCAAGTCGTAGTTCCACCATCAATCATCGATAATATGACGTCAACGGATATGGTGGTCCGGGAGAGTTCAGACGTGACCTTAGTTTGTCGTGCATCCGGGTATCCGGAGCCTTACGTCATGTGGAGGAGGGAAGATGGACAAGATTTCACTTACAACGGAGAATCAGTAAGTGTTGTTGACGGAGAAACGTTGACAATAACGAAGGTGTCTCGATTACATATGGGCGCCTACCTGTGCATCGCCTCTAATGGCGTGCCACCTTCAATTAGCAAGAGAATTATGCTCATGGTACAATTTCCGCCGATGCTTTCGATTCCCAATCAACTGGAAGGAGCGTACATAGGCCAGGATGTAACGTTAGAATGCCACACAGAAGCGTATCCATCTTCAATTAACTATTGGACAACAGATCGAGGGGATATGATCATTTCTGGTGGAAAATACGAAGCGTTCCCGATGGACAACGGTTACAGCAAATTTATGATGCTTAAAATACGAAACATAACTAAGGAAGATTTTGGATTCTACAAATGTATTGCAAAGAATTCACTAGGAGAGACCGATGGCATCATTAAGCTGGATGAAATTCCAGCGCCGCCGTCAGCGTTTACAACACAAAGAACTGTGTTAGATAATCAAACAATAAGGAAAAAAGGCGATGCACAAATCATTCAAAACAGTGACGCAATCACTTCAAATTTTCAAGGGGAGGCTTATGGAGAGCAAATTAGGGACTTTGATTTTGATGACGACGACAGCGCAAGCAACGATCTCCATTCAAACAGCCTATTATTTACCCTAATAGTTGGACTTGTATTACTCAAAGTTTTTAGGCAAAGTATACTTTCTTGA
Protein
MSKYGRELPSGQNVVSVAPIEVPDESMYPRFAEPIPNVTVTIGRDALLACVVENLRGFKVAWVRMDTQTILSIHHNIITQNSRISLSYNDHRSWYLHIKNVQEVDRGWYMCQVNTDPMHFRKGYLQVVVPPSIIDNMTSTDMVVRESSDVTLVCRASGYPEPYVMWRREDGQDFTYNGESVSVVDGETLTITKVSRLHMGAYLCIASNGVPPSISKRIMLMVQFPPMLSIPNQLEGAYIGQDVTLECHTEAYPSSINYWTTDRGDMIISGGKYEAFPMDNGYSKFMMLKIRNITKEDFGFYKCIAKNSLGETDGIIKLDEIPAPPSAFTTQRTVLDNQTIRKKGDAQIIQNSDAITSNFQGEAYGEQIRDFDFDDDDSASNDLHSNSLLFTLIVGLVLLKVFRQSILS
Summary
Uniprot
A0A194Q304
A0A194RTG6
H9JAF8
A0A146LH66
A0A023F4V5
A0A2A4J3T6
+ More
A0A146MA51 A0A3Q0J8E8 A0A139WMP7 A0A1B6CRI0 A0A1B6J1W7 A0A212EKA1 D6W8T8 H9JA27 A0A139WMS5 A0A3L8E4Q5 A0A026WAB6 A0A0A9XI90 A0A0K8T337 A0A146KK58 A0A151WZH5 A0A2S2QH63 A0A195BW86 J9JZN3 A0A0K8T3P7 A0A0A9XQI0 A0A195EIX3 A0A195D642 A0A146MGR1 A0A158P395 A0A2S2NNN0 A0A1S4GZZ7 A0A195F5V6 A0A088AQT0 B4KIR3 B4JP63 A0A154P4H3 B4N0B5 A0A2H8TG58 A0A1I8M262 A0A0C9RGF3 B4M9Q9 A0A1A9X2A9 A0A1A9ZHU6 A0A336LZ41 A0A0P9A7F1 A0A0R3NQJ6 A0A1W4V7R8 X2J5Z1 A0A0J9R1K5 B3MV45 A0A1A9X8Z5 Q29PB2 A0A3B0JIW5 A0A0C9QXP8 A0A1W4V797 B4Q545 A0A0R1DKD6 B3NAE9 B4IEP8 Q8IP70 B4P3N8 K7J2A2 A0A1B0FMB0 A0A232ET04 A0A1J1I230 A0A182KWN5 A0A2J7QVC7 A0A2S2QG06 A0A2S2N9P3 A0A2H8TP39 A0A2H8TEI3 A0A0P5NUT0 A0A164NFU9 A0A0P5R0R2 A0A0N8E0E1 A0A0P6AMJ1 E9H1U1 A0A0P6EB83 A0A182VSY0 A0A139WMQ5 A0A084WD23 A0A182N8R0 D6W8U0 A0A1S4H3E1 A0A182QF62
A0A146MA51 A0A3Q0J8E8 A0A139WMP7 A0A1B6CRI0 A0A1B6J1W7 A0A212EKA1 D6W8T8 H9JA27 A0A139WMS5 A0A3L8E4Q5 A0A026WAB6 A0A0A9XI90 A0A0K8T337 A0A146KK58 A0A151WZH5 A0A2S2QH63 A0A195BW86 J9JZN3 A0A0K8T3P7 A0A0A9XQI0 A0A195EIX3 A0A195D642 A0A146MGR1 A0A158P395 A0A2S2NNN0 A0A1S4GZZ7 A0A195F5V6 A0A088AQT0 B4KIR3 B4JP63 A0A154P4H3 B4N0B5 A0A2H8TG58 A0A1I8M262 A0A0C9RGF3 B4M9Q9 A0A1A9X2A9 A0A1A9ZHU6 A0A336LZ41 A0A0P9A7F1 A0A0R3NQJ6 A0A1W4V7R8 X2J5Z1 A0A0J9R1K5 B3MV45 A0A1A9X8Z5 Q29PB2 A0A3B0JIW5 A0A0C9QXP8 A0A1W4V797 B4Q545 A0A0R1DKD6 B3NAE9 B4IEP8 Q8IP70 B4P3N8 K7J2A2 A0A1B0FMB0 A0A232ET04 A0A1J1I230 A0A182KWN5 A0A2J7QVC7 A0A2S2QG06 A0A2S2N9P3 A0A2H8TP39 A0A2H8TEI3 A0A0P5NUT0 A0A164NFU9 A0A0P5R0R2 A0A0N8E0E1 A0A0P6AMJ1 E9H1U1 A0A0P6EB83 A0A182VSY0 A0A139WMQ5 A0A084WD23 A0A182N8R0 D6W8U0 A0A1S4H3E1 A0A182QF62
Pubmed
26354079
19121390
26823975
25474469
18362917
19820115
+ More
22118469 30249741 24508170 25401762 21347285 12364791 17994087 25315136 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 17550304 18057021 26109357 26109356 20075255 28648823 20966253 21292972 24438588
22118469 30249741 24508170 25401762 21347285 12364791 17994087 25315136 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 17550304 18057021 26109357 26109356 20075255 28648823 20966253 21292972 24438588
EMBL
KQ459551
KPI99921.1
KQ459896
KPJ19411.1
BABH01009272
GDHC01011924
+ More
JAQ06705.1 GBBI01002206 JAC16506.1 NWSH01003607 PCG66063.1 GDHC01002707 JAQ15922.1 KQ971312 KYB29126.1 GEDC01021485 JAS15813.1 GECU01014531 JAS93175.1 AGBW02014276 OWR41942.1 EEZ98396.2 BABH01009265 KYB29125.1 QOIP01000001 RLU27443.1 KK107304 EZA52923.1 GBHO01024243 GBRD01006009 JAG19361.1 JAG59812.1 GBRD01006010 JAG59811.1 GDHC01021648 JAP96980.1 KQ982649 KYQ53167.1 GGMS01007863 MBY77066.1 KQ976403 KYM92226.1 ABLF02040116 ABLF02040120 GBRD01006011 JAG59810.1 GBHO01024244 GBRD01006008 JAG19360.1 JAG59813.1 KQ978801 KYN28203.1 KQ976842 KYN07899.1 GDHC01000502 JAQ18127.1 ADTU01007920 ADTU01007921 ADTU01007922 ADTU01007923 ADTU01007924 ADTU01007925 ADTU01007926 ADTU01007927 ADTU01007928 GGMR01006181 MBY18800.1 AAAB01008944 KQ981805 KYN35454.1 CH933807 EDW12419.2 CH916372 EDV99488.1 KQ434804 KZC06020.1 CH963920 EDW77528.2 GFXV01001260 MBW13065.1 GBYB01007435 JAG77202.1 CH940654 EDW57935.2 UFQT01000047 SSX18948.1 CH902624 KPU74306.1 CH379058 KRT03441.1 AE014134 AHN54418.1 CM002910 KMY90142.1 EDV33110.1 EAL34381.2 OUUW01000006 SPP82317.1 GBYB01005452 JAG75219.1 CM000361 EDX04951.1 KMY90141.1 CM000157 KRJ97706.1 CH954177 EDV58651.1 KQS70467.1 KQS70468.1 CH480831 EDW46075.1 BT010243 AAN10840.1 AAQ23561.1 EDW88339.1 KRJ97704.1 KRJ97705.1 KRJ97707.1 KRJ97708.1 AAZX01001585 AAZX01004052 AAZX01008048 CCAG010002011 CCAG010002012 CCAG010002013 NNAY01002356 OXU21481.1 CVRI01000038 CRK94267.1 NEVH01010477 PNF32521.1 GGMS01007482 MBY76685.1 GGMR01001043 MBY13662.1 GFXV01004110 MBW15915.1 GFXV01000712 MBW12517.1 GDIQ01137430 JAL14296.1 LRGB01002860 KZS05919.1 GDIQ01107432 JAL44294.1 GDIQ01074140 JAN20597.1 GDIP01027146 JAM76569.1 GL732584 EFX74233.1 GDIQ01065249 JAN29488.1 KYB29127.1 ATLV01022922 ATLV01022923 KE525338 KFB48117.1 EEZ98398.2 AAAB01008980 AXCN02001860 AXCN02001861
JAQ06705.1 GBBI01002206 JAC16506.1 NWSH01003607 PCG66063.1 GDHC01002707 JAQ15922.1 KQ971312 KYB29126.1 GEDC01021485 JAS15813.1 GECU01014531 JAS93175.1 AGBW02014276 OWR41942.1 EEZ98396.2 BABH01009265 KYB29125.1 QOIP01000001 RLU27443.1 KK107304 EZA52923.1 GBHO01024243 GBRD01006009 JAG19361.1 JAG59812.1 GBRD01006010 JAG59811.1 GDHC01021648 JAP96980.1 KQ982649 KYQ53167.1 GGMS01007863 MBY77066.1 KQ976403 KYM92226.1 ABLF02040116 ABLF02040120 GBRD01006011 JAG59810.1 GBHO01024244 GBRD01006008 JAG19360.1 JAG59813.1 KQ978801 KYN28203.1 KQ976842 KYN07899.1 GDHC01000502 JAQ18127.1 ADTU01007920 ADTU01007921 ADTU01007922 ADTU01007923 ADTU01007924 ADTU01007925 ADTU01007926 ADTU01007927 ADTU01007928 GGMR01006181 MBY18800.1 AAAB01008944 KQ981805 KYN35454.1 CH933807 EDW12419.2 CH916372 EDV99488.1 KQ434804 KZC06020.1 CH963920 EDW77528.2 GFXV01001260 MBW13065.1 GBYB01007435 JAG77202.1 CH940654 EDW57935.2 UFQT01000047 SSX18948.1 CH902624 KPU74306.1 CH379058 KRT03441.1 AE014134 AHN54418.1 CM002910 KMY90142.1 EDV33110.1 EAL34381.2 OUUW01000006 SPP82317.1 GBYB01005452 JAG75219.1 CM000361 EDX04951.1 KMY90141.1 CM000157 KRJ97706.1 CH954177 EDV58651.1 KQS70467.1 KQS70468.1 CH480831 EDW46075.1 BT010243 AAN10840.1 AAQ23561.1 EDW88339.1 KRJ97704.1 KRJ97705.1 KRJ97707.1 KRJ97708.1 AAZX01001585 AAZX01004052 AAZX01008048 CCAG010002011 CCAG010002012 CCAG010002013 NNAY01002356 OXU21481.1 CVRI01000038 CRK94267.1 NEVH01010477 PNF32521.1 GGMS01007482 MBY76685.1 GGMR01001043 MBY13662.1 GFXV01004110 MBW15915.1 GFXV01000712 MBW12517.1 GDIQ01137430 JAL14296.1 LRGB01002860 KZS05919.1 GDIQ01107432 JAL44294.1 GDIQ01074140 JAN20597.1 GDIP01027146 JAM76569.1 GL732584 EFX74233.1 GDIQ01065249 JAN29488.1 KYB29127.1 ATLV01022922 ATLV01022923 KE525338 KFB48117.1 EEZ98398.2 AAAB01008980 AXCN02001860 AXCN02001861
Proteomes
UP000053268
UP000053240
UP000005204
UP000218220
UP000079169
UP000007266
+ More
UP000007151 UP000279307 UP000053097 UP000075809 UP000078540 UP000007819 UP000078492 UP000078542 UP000005205 UP000078541 UP000005203 UP000009192 UP000001070 UP000076502 UP000007798 UP000095301 UP000008792 UP000091820 UP000092445 UP000007801 UP000001819 UP000192221 UP000000803 UP000092443 UP000268350 UP000000304 UP000002282 UP000008711 UP000001292 UP000002358 UP000092444 UP000215335 UP000183832 UP000075882 UP000235965 UP000076858 UP000000305 UP000075920 UP000030765 UP000075884 UP000075886
UP000007151 UP000279307 UP000053097 UP000075809 UP000078540 UP000007819 UP000078492 UP000078542 UP000005205 UP000078541 UP000005203 UP000009192 UP000001070 UP000076502 UP000007798 UP000095301 UP000008792 UP000091820 UP000092445 UP000007801 UP000001819 UP000192221 UP000000803 UP000092443 UP000268350 UP000000304 UP000002282 UP000008711 UP000001292 UP000002358 UP000092444 UP000215335 UP000183832 UP000075882 UP000235965 UP000076858 UP000000305 UP000075920 UP000030765 UP000075884 UP000075886
Interpro
SUPFAM
SSF48726
SSF48726
Gene 3D
ProteinModelPortal
A0A194Q304
A0A194RTG6
H9JAF8
A0A146LH66
A0A023F4V5
A0A2A4J3T6
+ More
A0A146MA51 A0A3Q0J8E8 A0A139WMP7 A0A1B6CRI0 A0A1B6J1W7 A0A212EKA1 D6W8T8 H9JA27 A0A139WMS5 A0A3L8E4Q5 A0A026WAB6 A0A0A9XI90 A0A0K8T337 A0A146KK58 A0A151WZH5 A0A2S2QH63 A0A195BW86 J9JZN3 A0A0K8T3P7 A0A0A9XQI0 A0A195EIX3 A0A195D642 A0A146MGR1 A0A158P395 A0A2S2NNN0 A0A1S4GZZ7 A0A195F5V6 A0A088AQT0 B4KIR3 B4JP63 A0A154P4H3 B4N0B5 A0A2H8TG58 A0A1I8M262 A0A0C9RGF3 B4M9Q9 A0A1A9X2A9 A0A1A9ZHU6 A0A336LZ41 A0A0P9A7F1 A0A0R3NQJ6 A0A1W4V7R8 X2J5Z1 A0A0J9R1K5 B3MV45 A0A1A9X8Z5 Q29PB2 A0A3B0JIW5 A0A0C9QXP8 A0A1W4V797 B4Q545 A0A0R1DKD6 B3NAE9 B4IEP8 Q8IP70 B4P3N8 K7J2A2 A0A1B0FMB0 A0A232ET04 A0A1J1I230 A0A182KWN5 A0A2J7QVC7 A0A2S2QG06 A0A2S2N9P3 A0A2H8TP39 A0A2H8TEI3 A0A0P5NUT0 A0A164NFU9 A0A0P5R0R2 A0A0N8E0E1 A0A0P6AMJ1 E9H1U1 A0A0P6EB83 A0A182VSY0 A0A139WMQ5 A0A084WD23 A0A182N8R0 D6W8U0 A0A1S4H3E1 A0A182QF62
A0A146MA51 A0A3Q0J8E8 A0A139WMP7 A0A1B6CRI0 A0A1B6J1W7 A0A212EKA1 D6W8T8 H9JA27 A0A139WMS5 A0A3L8E4Q5 A0A026WAB6 A0A0A9XI90 A0A0K8T337 A0A146KK58 A0A151WZH5 A0A2S2QH63 A0A195BW86 J9JZN3 A0A0K8T3P7 A0A0A9XQI0 A0A195EIX3 A0A195D642 A0A146MGR1 A0A158P395 A0A2S2NNN0 A0A1S4GZZ7 A0A195F5V6 A0A088AQT0 B4KIR3 B4JP63 A0A154P4H3 B4N0B5 A0A2H8TG58 A0A1I8M262 A0A0C9RGF3 B4M9Q9 A0A1A9X2A9 A0A1A9ZHU6 A0A336LZ41 A0A0P9A7F1 A0A0R3NQJ6 A0A1W4V7R8 X2J5Z1 A0A0J9R1K5 B3MV45 A0A1A9X8Z5 Q29PB2 A0A3B0JIW5 A0A0C9QXP8 A0A1W4V797 B4Q545 A0A0R1DKD6 B3NAE9 B4IEP8 Q8IP70 B4P3N8 K7J2A2 A0A1B0FMB0 A0A232ET04 A0A1J1I230 A0A182KWN5 A0A2J7QVC7 A0A2S2QG06 A0A2S2N9P3 A0A2H8TP39 A0A2H8TEI3 A0A0P5NUT0 A0A164NFU9 A0A0P5R0R2 A0A0N8E0E1 A0A0P6AMJ1 E9H1U1 A0A0P6EB83 A0A182VSY0 A0A139WMQ5 A0A084WD23 A0A182N8R0 D6W8U0 A0A1S4H3E1 A0A182QF62
PDB
6EG0
E-value=1.26211e-98,
Score=919
Ontologies
GO
Topology
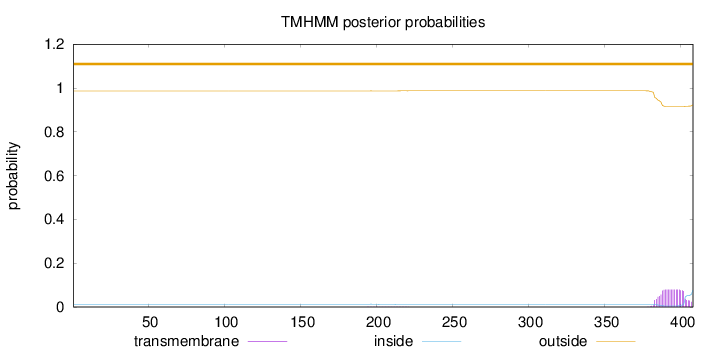
Length:
408
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.58728
Exp number, first 60 AAs:
0.00122
Total prob of N-in:
0.01306
outside
1 - 408
Population Genetic Test Statistics
Pi
222.007007
Theta
170.528818
Tajima's D
0.930036
CLR
0.000005
CSRT
0.643717814109295
Interpretation
Uncertain
