Gene
KWMTBOMO03415
Pre Gene Modal
BGIBMGA006374
Annotation
PREDICTED:_RUN_and_FYVE_domain-containing_protein_2_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.833
Sequence
CDS
ATGGCATCTGCAAGAGAAAATGATGCAGAAGTATCACCAACATCATCTATGTCTTCTGTCAAATCTCATCCCATCGAAAGATCTTCATCTTCAAAAGATGAAAGATGGCCAGAACTTCTGATATCGCGACCACAAAAAACATATAACTCAAAAACTCGCGATCCCGTTGTCATTGAGCGCAGTAATCTCGTGAATATCTCGAAACTTATAGTGAAGGAACTAATCGAACAGTCGCTTCGTTTCGGCCGAATGCTCGACAGTGATCACTTGCCGTTGCATCACTTCTTTATTGTCCTCGAACATGTCATGCGTCATGGACTCAAACCGAAAAAGGGACTCCTCGGACCTAAAAAGGAGCTATGGGATATTCTTCAAATGGTTGAGAAGTATTCGCCTGAGGCAGCCGATATCACGGCCAGTGTCAGAGATCTTCCAACTGTGAAGACAGCAATGGGCCGCGCTCGGGCTTGGTTGCGATTGGCGTTAATGCAAAAACGACTCGCAGATTACTTGAGAATCCTCCTTGAGCATCGTGAAGATACTTTGGAGGAATATTTCGAACCCCACGCGCTGATGCTGAACGAGGAAGCTGTAATTATCACCGGTTTGCTGGTCGGTCTCAATGTTGTCGACTGCAATTTGTGTGTTAAGGAAGAAGACTTAGATAGTCAACAAGGTGTCATAGATTTCTCTTTGTATTTACGGGGCAGTAACTCTTCCAACGACATAGCAGGAAATGAGAACAACACAAACAATGAACCTAAACAACGCCACATCAACACCATGCTCGATCAGAAAAACTATATTGAAGAACTAAACCGACATCTGAACGCCACCGTAGCAAATCTCCAGGCGAAAGTCGAAAGCCTGACCACGACCAATGCGTTGATGAAGGAAGACCTTGCAATCGCAAAGAACACAATGATTCAGCTAGTCGAAGAAAACAATCAACTGAAACACGCTTTAGGTCAAGATATAACCAAAGATTCGAGAATGAAAATCACGGAACTGCAAGCAGATGAAGAGGAAAAACGAAGCGTTAAAAGTGTAACTTCTGATAGATCTAAGAGTGATAAGGATAGCGAGATGTCAAAGGCGTTAGAGGAAGAAAGGAAGAAGTATTTAGAAATGCAAAAGGAACTTGATTTGCAGATTTCATTGAAGGCAGAAATGGAGGTAGCCATGAAATTATTAGAAAAAGATATACACGACAAACAAGACACTATTATATCTCTGAGGAGGCAGCTTGACGATATAAAGCTTATCAATTTAGAAATGTACAAAAAATTACAGGAATGTGAAATTTCCTTGAAACATAAAACAGAGTTAATAGCCAAATTGGAAGCAAAGACTGAATCAATGGGCAGCACCATACATCAACTCGATGAAAAGTTACAAGAAGATAAACTAACTAGGAAGAATCTAGAAGATAATGCCCGCACATTGGGTCAAAAAGCGACCACAGCCGAAAAGAAAGCTGTGGCAGCAGAGGGAGACCTTAGAATTGAAAGAGAATGGAGGATATCTCTACAAGAATCTATGATGCGAGACCGAGACAAAATATCAAGTCTTACACAAGAAGTTGAGTCTTTGAAAGCCATTGGACAGAAATATCTTCTTTTACAAGAAGAGCAACACATCTTGAAGACTAAATACAGCGAAGCACAAAAAACTTTAGAAGAAGTCGGAGCCACGCTCAGCGAGAACAAACTTCAGCTAGCTGAGTTGTTAGAGAAGGAATCATTAGCAACTACCACAGAGGAAACACCCACTTGGGCCAACGATAAAGACGCGACGGCTTGCGCTACGTGTAATAAGGAATTCAATATTACAAGACGGAAACATCACTGTAGAATGTGTGGTCTAATATTTTGCGCAGCTTGCAGTGATAAAACTGTTGCATTGACTGGCAACACCAAACCTGTTCGTGTTTGTGGTGCGTGCTACACGGAAGCTGTCCGCCACACAGAGTAA
Protein
MASARENDAEVSPTSSMSSVKSHPIERSSSSKDERWPELLISRPQKTYNSKTRDPVVIERSNLVNISKLIVKELIEQSLRFGRMLDSDHLPLHHFFIVLEHVMRHGLKPKKGLLGPKKELWDILQMVEKYSPEAADITASVRDLPTVKTAMGRARAWLRLALMQKRLADYLRILLEHREDTLEEYFEPHALMLNEEAVIITGLLVGLNVVDCNLCVKEEDLDSQQGVIDFSLYLRGSNSSNDIAGNENNTNNEPKQRHINTMLDQKNYIEELNRHLNATVANLQAKVESLTTTNALMKEDLAIAKNTMIQLVEENNQLKHALGQDITKDSRMKITELQADEEEKRSVKSVTSDRSKSDKDSEMSKALEEERKKYLEMQKELDLQISLKAEMEVAMKLLEKDIHDKQDTIISLRRQLDDIKLINLEMYKKLQECEISLKHKTELIAKLEAKTESMGSTIHQLDEKLQEDKLTRKNLEDNARTLGQKATTAEKKAVAAEGDLRIEREWRISLQESMMRDRDKISSLTQEVESLKAIGQKYLLLQEEQHILKTKYSEAQKTLEEVGATLSENKLQLAELLEKESLATTTEETPTWANDKDATACATCNKEFNITRRKHHCRMCGLIFCAACSDKTVALTGNTKPVRVCGACYTEAVRHTE
Summary
Uniprot
A0A1E1WP88
A0A194Q4I7
A0A2A4JUY1
A0A1E1WNZ6
A0A1E1W2Q1
A0A067QY49
+ More
A0A2J7Q478 A0A026WD06 A0A2J7Q461 A0A151WZP3 A0A3L8E143 A0A0C9QHC8 A0A195F4Z1 A0A195EK18 A0A1W4XF33 A0A1B6KDL4 A0A087ZRI3 A0A1W4X4L6 A0A1S4E6A5 A0A158P397 A0A1S4F1N7 A0A1L8DMD8 A0A1Q3FUB2 A0A0K8TM47 A0A0Q9WU44 A0A0A9Z5E2 A0A0K8TCQ7 W8B686 A0A0Q9X263 A0A023F3I7 A0A0Q9X1T5 A0A0A9XDF2 B4K5Z8 A0A336MP46 A0A0Q9X1S0 A0A336MTN5 A0A069DVU9 A0A1S4E6B6 A0A336MRG5 A0A1B6DMN5 A0A182F8M0 A0A182R826 A0A2M4AGZ8 A0A2M4AGV5 A0A2M4AGZ0 A0A1I8Q9M0 A0A1I8Q9N5 A0A0B4LHR8 A0A1Y9HBV3 A0A0P4VSM2 A0A084VI68 A0A182JCH9 A0A1W4URN7 A0A340TB50 A0A1I8Q9R8 Q8IMP2 F5HK19 A0A1B6EEQ6 A0A0K8V3G7 A0A1I8NGL8 A0A1I8NGL5 B4PRT7 A0A0R1E544 A0A0P8YGQ7 A0A0P8Y1A8 A0A034VGG0 A0A0N8P1Q0 A0A0A1X5B7 B4M5R6 A0A0Q9W0H9 A0A3B0JFU8 I5ANI8 A0A0R3NLS0 T1IKM3 A0A2R5LMI0 L7MCR1 A0A0L0C7I2 A0A1J1I8F7 A0A087UC24 Q17IA1 A0A1E1XHK3 A0A131XCI8 A0A2L2YQS6 A0A195D4V6 J9JSV1 A0A1S3I786 A0A2T7Q1M3 A0A1S3I7A0 A0A2R2MRA7 A0A2R2MSA4 E9G458 A0A0K2T5U1 A0A0T6B3A8
A0A2J7Q478 A0A026WD06 A0A2J7Q461 A0A151WZP3 A0A3L8E143 A0A0C9QHC8 A0A195F4Z1 A0A195EK18 A0A1W4XF33 A0A1B6KDL4 A0A087ZRI3 A0A1W4X4L6 A0A1S4E6A5 A0A158P397 A0A1S4F1N7 A0A1L8DMD8 A0A1Q3FUB2 A0A0K8TM47 A0A0Q9WU44 A0A0A9Z5E2 A0A0K8TCQ7 W8B686 A0A0Q9X263 A0A023F3I7 A0A0Q9X1T5 A0A0A9XDF2 B4K5Z8 A0A336MP46 A0A0Q9X1S0 A0A336MTN5 A0A069DVU9 A0A1S4E6B6 A0A336MRG5 A0A1B6DMN5 A0A182F8M0 A0A182R826 A0A2M4AGZ8 A0A2M4AGV5 A0A2M4AGZ0 A0A1I8Q9M0 A0A1I8Q9N5 A0A0B4LHR8 A0A1Y9HBV3 A0A0P4VSM2 A0A084VI68 A0A182JCH9 A0A1W4URN7 A0A340TB50 A0A1I8Q9R8 Q8IMP2 F5HK19 A0A1B6EEQ6 A0A0K8V3G7 A0A1I8NGL8 A0A1I8NGL5 B4PRT7 A0A0R1E544 A0A0P8YGQ7 A0A0P8Y1A8 A0A034VGG0 A0A0N8P1Q0 A0A0A1X5B7 B4M5R6 A0A0Q9W0H9 A0A3B0JFU8 I5ANI8 A0A0R3NLS0 T1IKM3 A0A2R5LMI0 L7MCR1 A0A0L0C7I2 A0A1J1I8F7 A0A087UC24 Q17IA1 A0A1E1XHK3 A0A131XCI8 A0A2L2YQS6 A0A195D4V6 J9JSV1 A0A1S3I786 A0A2T7Q1M3 A0A1S3I7A0 A0A2R2MRA7 A0A2R2MSA4 E9G458 A0A0K2T5U1 A0A0T6B3A8
Pubmed
26354079
24845553
24508170
30249741
21347285
26369729
+ More
17994087 25401762 24495485 25474469 18057021 26334808 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 27129103 24438588 26109357 26109356 12364791 14747013 17210077 25315136 17550304 25348373 25830018 15632085 25576852 26108605 17510324 28503490 28049606 26561354 21292972
17994087 25401762 24495485 25474469 18057021 26334808 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 27129103 24438588 26109357 26109356 12364791 14747013 17210077 25315136 17550304 25348373 25830018 15632085 25576852 26108605 17510324 28503490 28049606 26561354 21292972
EMBL
GDQN01002403
JAT88651.1
KQ459551
KPI99924.1
NWSH01000532
PCG75835.1
+ More
GDQN01002339 JAT88715.1 GDQN01009873 GDQN01007381 JAT81181.1 JAT83673.1 KK853216 KDR09775.1 NEVH01018390 PNF23375.1 KK107304 EZA52929.1 PNF23373.1 KQ982649 KYQ53171.1 QOIP01000001 RLU26346.1 GBYB01014038 JAG83805.1 KQ981805 KYN35461.1 KQ978801 KYN28209.1 GEBQ01030429 JAT09548.1 ADTU01007944 ADTU01007945 ADTU01007946 ADTU01007947 GFDF01006584 JAV07500.1 GFDL01003917 JAV31128.1 GDAI01002598 JAI15005.1 CH964251 KRF99695.1 GBHO01003930 JAG39674.1 GBRD01002630 JAG63191.1 GAMC01017729 JAB88826.1 CH933806 KRG02013.1 GBBI01002779 JAC15933.1 KRG02010.1 KRG02011.1 GBHO01025580 GBRD01002631 JAG18024.1 JAG63190.1 EDW16235.2 UFQT01001912 SSX32142.1 KRG02015.1 SSX32143.1 GBGD01000943 JAC87946.1 SSX32141.1 GEDC01010408 JAS26890.1 GGFK01006739 MBW40060.1 GGFK01006702 MBW40023.1 GGFK01006738 MBW40059.1 AE014297 AHN57559.1 GDKW01002620 JAI53975.1 ATLV01013301 KE524850 KFB37662.1 BT044470 AAN14401.1 ACH92535.1 AAAB01008851 EGK96630.1 GEDC01000890 JAS36408.1 GDHF01018870 GDHF01011926 JAI33444.1 JAI40388.1 CM000160 EDW98530.2 KRK04328.1 CH902617 KPU80539.1 KPU80543.1 GAKP01018087 JAC40865.1 KPU80540.1 GBXI01007763 JAD06529.1 CH940652 EDW58992.2 KRF78636.1 KRF78634.1 OUUW01000005 SPP80995.1 CM000070 EIM52523.1 KRT00080.1 JH430577 GGLE01006617 MBY10743.1 GACK01003447 JAA61587.1 JRES01000809 KNC28217.1 CVRI01000044 CRK96552.1 KK119164 KFM74913.1 CH477241 EAT46394.1 GFAC01000453 JAT98735.1 GEFH01004489 JAP64092.1 IAAA01040096 LAA10367.1 KQ976842 KYN07907.1 ABLF02035465 PZQS01000001 PVD39571.1 GL732531 EFX85697.1 HACA01003824 CDW21185.1 LJIG01016056 KRT81748.1
GDQN01002339 JAT88715.1 GDQN01009873 GDQN01007381 JAT81181.1 JAT83673.1 KK853216 KDR09775.1 NEVH01018390 PNF23375.1 KK107304 EZA52929.1 PNF23373.1 KQ982649 KYQ53171.1 QOIP01000001 RLU26346.1 GBYB01014038 JAG83805.1 KQ981805 KYN35461.1 KQ978801 KYN28209.1 GEBQ01030429 JAT09548.1 ADTU01007944 ADTU01007945 ADTU01007946 ADTU01007947 GFDF01006584 JAV07500.1 GFDL01003917 JAV31128.1 GDAI01002598 JAI15005.1 CH964251 KRF99695.1 GBHO01003930 JAG39674.1 GBRD01002630 JAG63191.1 GAMC01017729 JAB88826.1 CH933806 KRG02013.1 GBBI01002779 JAC15933.1 KRG02010.1 KRG02011.1 GBHO01025580 GBRD01002631 JAG18024.1 JAG63190.1 EDW16235.2 UFQT01001912 SSX32142.1 KRG02015.1 SSX32143.1 GBGD01000943 JAC87946.1 SSX32141.1 GEDC01010408 JAS26890.1 GGFK01006739 MBW40060.1 GGFK01006702 MBW40023.1 GGFK01006738 MBW40059.1 AE014297 AHN57559.1 GDKW01002620 JAI53975.1 ATLV01013301 KE524850 KFB37662.1 BT044470 AAN14401.1 ACH92535.1 AAAB01008851 EGK96630.1 GEDC01000890 JAS36408.1 GDHF01018870 GDHF01011926 JAI33444.1 JAI40388.1 CM000160 EDW98530.2 KRK04328.1 CH902617 KPU80539.1 KPU80543.1 GAKP01018087 JAC40865.1 KPU80540.1 GBXI01007763 JAD06529.1 CH940652 EDW58992.2 KRF78636.1 KRF78634.1 OUUW01000005 SPP80995.1 CM000070 EIM52523.1 KRT00080.1 JH430577 GGLE01006617 MBY10743.1 GACK01003447 JAA61587.1 JRES01000809 KNC28217.1 CVRI01000044 CRK96552.1 KK119164 KFM74913.1 CH477241 EAT46394.1 GFAC01000453 JAT98735.1 GEFH01004489 JAP64092.1 IAAA01040096 LAA10367.1 KQ976842 KYN07907.1 ABLF02035465 PZQS01000001 PVD39571.1 GL732531 EFX85697.1 HACA01003824 CDW21185.1 LJIG01016056 KRT81748.1
Proteomes
UP000053268
UP000218220
UP000027135
UP000235965
UP000053097
UP000075809
+ More
UP000279307 UP000078541 UP000078492 UP000192223 UP000005203 UP000079169 UP000005205 UP000007798 UP000009192 UP000069272 UP000075900 UP000095300 UP000000803 UP000030765 UP000075880 UP000192221 UP000075903 UP000007062 UP000095301 UP000002282 UP000007801 UP000008792 UP000268350 UP000001819 UP000037069 UP000183832 UP000054359 UP000008820 UP000078542 UP000007819 UP000085678 UP000245119 UP000000305
UP000279307 UP000078541 UP000078492 UP000192223 UP000005203 UP000079169 UP000005205 UP000007798 UP000009192 UP000069272 UP000075900 UP000095300 UP000000803 UP000030765 UP000075880 UP000192221 UP000075903 UP000007062 UP000095301 UP000002282 UP000007801 UP000008792 UP000268350 UP000001819 UP000037069 UP000183832 UP000054359 UP000008820 UP000078542 UP000007819 UP000085678 UP000245119 UP000000305
PRIDE
Interpro
IPR037213
Run_dom_sf
+ More
IPR000306 Znf_FYVE
IPR001841 Znf_RING
IPR004012 Run_dom
IPR011011 Znf_FYVE_PHD
IPR017455 Znf_FYVE-rel
IPR013083 Znf_RING/FYVE/PHD
IPR020861 Triosephosphate_isomerase_AS
IPR013785 Aldolase_TIM
IPR022896 TrioseP_Isoase_bac/euk
IPR000652 Triosephosphate_isomerase
IPR035990 TIM_sf
IPR000306 Znf_FYVE
IPR001841 Znf_RING
IPR004012 Run_dom
IPR011011 Znf_FYVE_PHD
IPR017455 Znf_FYVE-rel
IPR013083 Znf_RING/FYVE/PHD
IPR020861 Triosephosphate_isomerase_AS
IPR013785 Aldolase_TIM
IPR022896 TrioseP_Isoase_bac/euk
IPR000652 Triosephosphate_isomerase
IPR035990 TIM_sf
Gene 3D
CDD
ProteinModelPortal
A0A1E1WP88
A0A194Q4I7
A0A2A4JUY1
A0A1E1WNZ6
A0A1E1W2Q1
A0A067QY49
+ More
A0A2J7Q478 A0A026WD06 A0A2J7Q461 A0A151WZP3 A0A3L8E143 A0A0C9QHC8 A0A195F4Z1 A0A195EK18 A0A1W4XF33 A0A1B6KDL4 A0A087ZRI3 A0A1W4X4L6 A0A1S4E6A5 A0A158P397 A0A1S4F1N7 A0A1L8DMD8 A0A1Q3FUB2 A0A0K8TM47 A0A0Q9WU44 A0A0A9Z5E2 A0A0K8TCQ7 W8B686 A0A0Q9X263 A0A023F3I7 A0A0Q9X1T5 A0A0A9XDF2 B4K5Z8 A0A336MP46 A0A0Q9X1S0 A0A336MTN5 A0A069DVU9 A0A1S4E6B6 A0A336MRG5 A0A1B6DMN5 A0A182F8M0 A0A182R826 A0A2M4AGZ8 A0A2M4AGV5 A0A2M4AGZ0 A0A1I8Q9M0 A0A1I8Q9N5 A0A0B4LHR8 A0A1Y9HBV3 A0A0P4VSM2 A0A084VI68 A0A182JCH9 A0A1W4URN7 A0A340TB50 A0A1I8Q9R8 Q8IMP2 F5HK19 A0A1B6EEQ6 A0A0K8V3G7 A0A1I8NGL8 A0A1I8NGL5 B4PRT7 A0A0R1E544 A0A0P8YGQ7 A0A0P8Y1A8 A0A034VGG0 A0A0N8P1Q0 A0A0A1X5B7 B4M5R6 A0A0Q9W0H9 A0A3B0JFU8 I5ANI8 A0A0R3NLS0 T1IKM3 A0A2R5LMI0 L7MCR1 A0A0L0C7I2 A0A1J1I8F7 A0A087UC24 Q17IA1 A0A1E1XHK3 A0A131XCI8 A0A2L2YQS6 A0A195D4V6 J9JSV1 A0A1S3I786 A0A2T7Q1M3 A0A1S3I7A0 A0A2R2MRA7 A0A2R2MSA4 E9G458 A0A0K2T5U1 A0A0T6B3A8
A0A2J7Q478 A0A026WD06 A0A2J7Q461 A0A151WZP3 A0A3L8E143 A0A0C9QHC8 A0A195F4Z1 A0A195EK18 A0A1W4XF33 A0A1B6KDL4 A0A087ZRI3 A0A1W4X4L6 A0A1S4E6A5 A0A158P397 A0A1S4F1N7 A0A1L8DMD8 A0A1Q3FUB2 A0A0K8TM47 A0A0Q9WU44 A0A0A9Z5E2 A0A0K8TCQ7 W8B686 A0A0Q9X263 A0A023F3I7 A0A0Q9X1T5 A0A0A9XDF2 B4K5Z8 A0A336MP46 A0A0Q9X1S0 A0A336MTN5 A0A069DVU9 A0A1S4E6B6 A0A336MRG5 A0A1B6DMN5 A0A182F8M0 A0A182R826 A0A2M4AGZ8 A0A2M4AGV5 A0A2M4AGZ0 A0A1I8Q9M0 A0A1I8Q9N5 A0A0B4LHR8 A0A1Y9HBV3 A0A0P4VSM2 A0A084VI68 A0A182JCH9 A0A1W4URN7 A0A340TB50 A0A1I8Q9R8 Q8IMP2 F5HK19 A0A1B6EEQ6 A0A0K8V3G7 A0A1I8NGL8 A0A1I8NGL5 B4PRT7 A0A0R1E544 A0A0P8YGQ7 A0A0P8Y1A8 A0A034VGG0 A0A0N8P1Q0 A0A0A1X5B7 B4M5R6 A0A0Q9W0H9 A0A3B0JFU8 I5ANI8 A0A0R3NLS0 T1IKM3 A0A2R5LMI0 L7MCR1 A0A0L0C7I2 A0A1J1I8F7 A0A087UC24 Q17IA1 A0A1E1XHK3 A0A131XCI8 A0A2L2YQS6 A0A195D4V6 J9JSV1 A0A1S3I786 A0A2T7Q1M3 A0A1S3I7A0 A0A2R2MRA7 A0A2R2MSA4 E9G458 A0A0K2T5U1 A0A0T6B3A8
PDB
2CXL
E-value=5.67759e-52,
Score=518
Ontologies
GO
Topology
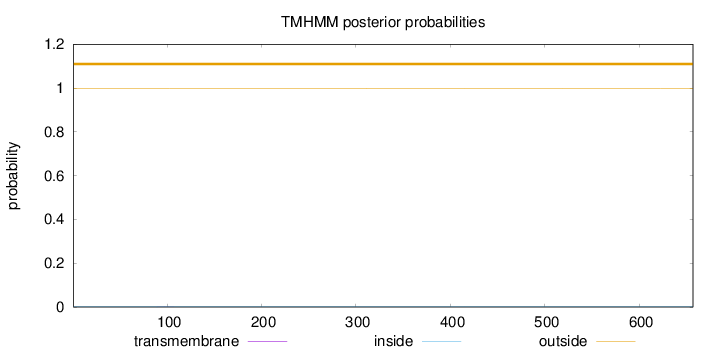
Length:
657
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0059
Exp number, first 60 AAs:
5e-05
Total prob of N-in:
0.00100
outside
1 - 657
Population Genetic Test Statistics
Pi
291.957775
Theta
201.24279
Tajima's D
1.331697
CLR
0.109809
CSRT
0.746062696865157
Interpretation
Uncertain
