Gene
KWMTBOMO03414
Pre Gene Modal
BGIBMGA006502
Annotation
Serine_carboxypeptidase-like_51_[Papilio_machaon]
Full name
Carboxypeptidase
+ More
Retinoid-inducible serine carboxypeptidase
Retinoid-inducible serine carboxypeptidase
Alternative Name
Serine carboxypeptidase 1
Location in the cell
Mitochondrial Reliability : 1.716 PlasmaMembrane Reliability : 1.073
Sequence
CDS
ATGTATCCTTCTACTCGACCACCTTTCTTCTCAGGAGGATACTTCGGTAATAATCCATATAGACCGCAACCGATACAACATCCATCTGACCATGTAAAACCTGTCCACGAAAGCAATAGCCAACAATTTCAAGAAATTTATATTCCAAATATTGTAGGTGTTATGATATCATCACGTTGTGCTGTGCGTTTAATTGTGGTTAATCGTTTTAGAGCGGTGCCAAAGCGCACATGTAAATTAGGAAGTGCTTGTGTATATTTTGTAAAACACACAATGCTAGTAAAATTATTTTTTTGTTTTTTCGCAACATTCTGCATTGCTAGCCAATCTGCACCTGAAGAAAATCCATATTTGCGGGCCGTGAATGAGAATTATTTAGCACAAAACTCCATACACGACTATGTAGAAGTTCGACCAGGTGCACACCTTTTTTATTGGTTTTTTTACGCGGACGGGACGAAGAAAGACGCATCTCGAAAGCCTCTTATAATATGGGTCCAAGGAGGTCCTGGTTTTGCCGCTAGTGGCGTAGGTAATTTTGGCGAATTCGGCCCCCTGACGATGGACATGCAACCAAGGAATCATACTTGGGTAAAAGGAAGGAATTTATTACTTATTGACCACCCCGTGGGCGCTGGATTTAGTTACGTCACAAGTAAAGATCTTTTTGTGAAAACAGATCAACAAATGGCTATTGACCTCTCACGAGCTATTAAACAATTTTTCAAAAAACACAAAGTTTTCCGAAAAACTCCTGCGTATGTGATTGGTCAGAGTTACGGCGGTAAATTGTGTCCAAGATTAGCTGTTTATTTATATACGGCTATCAAGAAAAAGAGTTTAAAAATGAACTTTAAAGGTATCGGTATTGGAGGTGGTTGGGTCAATCCTAAAGAGAGCACTTTAGTAACGCCACGGTTTCTTTATCATACCGGAGCAATCGATCGAGATTTATATCTGAGCTCTTCACGCATAGCTCACGAATTGATAAGTTTGATGGACAAAAACGATTATATTCAAGCACACGAAGTGGACTTTATGCTCTTCAAAAAGCTGAACGAAGACGAATTTCTAAACTTCAATAATATTTATGGTCTCAGCCCTTACCCGGCATTGGTCAAACTTAATCTCCATGTAAACCAATATGTTAAGCCGACATTAAAATTGGTGAATCAAACAATTGATTGGCTATACTTAAATACAGATGTTTATGATAGTTTAAAAGGAAGTCTTCTTGTTCCGTCTTTCTGGTTTTTGGAGAAAATTTTGAGCAAGACAAATTTGAAGGTTGTGATTTACAACGGAAACTTCGACGTGGTTACTCCATTAGCAGGTGCTTCAAACTGGGTTCACAAACTGAACTGGCCTCTTAAAGAGAATTTCACGAAGGCGGTACGACATCACATACGGGGCACAAATAATGGATATTATAAAGAAATGGAGCAACTTAGTTTTTGGTCTGTCTTTCGTGCCGGACATTGGGTTCCAGAAGATAACCCCGAAGCCATGGAACATATTTTAGAATATCTCCTTACTGAGAACAATTAG
Protein
MYPSTRPPFFSGGYFGNNPYRPQPIQHPSDHVKPVHESNSQQFQEIYIPNIVGVMISSRCAVRLIVVNRFRAVPKRTCKLGSACVYFVKHTMLVKLFFCFFATFCIASQSAPEENPYLRAVNENYLAQNSIHDYVEVRPGAHLFYWFFYADGTKKDASRKPLIIWVQGGPGFAASGVGNFGEFGPLTMDMQPRNHTWVKGRNLLLIDHPVGAGFSYVTSKDLFVKTDQQMAIDLSRAIKQFFKKHKVFRKTPAYVIGQSYGGKLCPRLAVYLYTAIKKKSLKMNFKGIGIGGGWVNPKESTLVTPRFLYHTGAIDRDLYLSSSRIAHELISLMDKNDYIQAHEVDFMLFKKLNEDEFLNFNNIYGLSPYPALVKLNLHVNQYVKPTLKLVNQTIDWLYLNTDVYDSLKGSLLVPSFWFLEKILSKTNLKVVIYNGNFDVVTPLAGASNWVHKLNWPLKENFTKAVRHHIRGTNNGYYKEMEQLSFWSVFRAGHWVPEDNPEAMEHILEYLLTENN
Summary
Similarity
Belongs to the peptidase S10 family.
Keywords
Carboxypeptidase
Complete proteome
Glycoprotein
Hydrolase
Protease
Reference proteome
Secreted
Signal
Feature
chain Carboxypeptidase
Uniprot
A0A2A4JWG7
A0A194RPH1
A0A2H1X335
A0A194Q928
A0A1W4XBY5
A0A1I8M8P6
+ More
B3N626 A0A1W4X145 A0A1I8NA52 B4NXT0 A0A0J9TP36 B4I1Y1 Q8IP31 A0A1L8EGA9 B3ML06 A0A0M4E2M3 A0A1W4VYE8 A0A1L8EFY1 A0A3G5BIH1 J3JW05 B4J2S3 D6W8Z8 B4N036 B4M8U4 U5EQW7 A0A3B0K9X1 A0A293MTR0 A0A0A1WN20 W8AS88 Q29KD6 B4H9E7 A0A1I8M708 A0A2L2Y5G2 A0A1V4KW23 A0A182GA02 B4L0B1 Q99J29 Q920A5 E0VVV2 A0A0C9PYP4 A0A1B0FRI3 A0A158P0S0 A0A2I0M7K2 B4JED0 A0A1A9VMJ8 A0A0M5J3G2 A0A1B0ASD3 K7IP64 A0A212ESD4 A0A1Y1MRG9 A0A1A9Y3F6 A0A1B0AEJ3 A0A1W7RAV1 R0M1S7 U3IBT5 A0A151N2K0 G3T5D9 A0A023GNA5 D6W901 B4KE97 A0A1I8PQA5 D6W8Z9 K7IP63 A0A2U3WA82 A0A1D5PBP3 B3MAE0 A0A1L8DPI1 A0A1U8DQI1 A0A0K8W077 K7J676 A0A2Y9LT36 A0A2U4C680 H9JLE7 N6UG27 H0ZH07 A0A091S5W8 A0A2P6L7L4 A0A341CHI8 A0A151WVZ7 A0A1L8DPK9 G1L7L9 A0A154NZ13 A0A2Y9JV52 A0A3L8SAC5 A0A091DS71 G3H1D5 A0A218UVF9 B4HVI6 A0A091IZ45 A0A2J6L2R3 A0A091HZT0 A0A0S3SCE7 A0A195CXH2 Q9W0N8 A0A232EW11 B4H4A7 Q29DW4 A0A226MSY2 F1PSP6 A0A094K4Z6
B3N626 A0A1W4X145 A0A1I8NA52 B4NXT0 A0A0J9TP36 B4I1Y1 Q8IP31 A0A1L8EGA9 B3ML06 A0A0M4E2M3 A0A1W4VYE8 A0A1L8EFY1 A0A3G5BIH1 J3JW05 B4J2S3 D6W8Z8 B4N036 B4M8U4 U5EQW7 A0A3B0K9X1 A0A293MTR0 A0A0A1WN20 W8AS88 Q29KD6 B4H9E7 A0A1I8M708 A0A2L2Y5G2 A0A1V4KW23 A0A182GA02 B4L0B1 Q99J29 Q920A5 E0VVV2 A0A0C9PYP4 A0A1B0FRI3 A0A158P0S0 A0A2I0M7K2 B4JED0 A0A1A9VMJ8 A0A0M5J3G2 A0A1B0ASD3 K7IP64 A0A212ESD4 A0A1Y1MRG9 A0A1A9Y3F6 A0A1B0AEJ3 A0A1W7RAV1 R0M1S7 U3IBT5 A0A151N2K0 G3T5D9 A0A023GNA5 D6W901 B4KE97 A0A1I8PQA5 D6W8Z9 K7IP63 A0A2U3WA82 A0A1D5PBP3 B3MAE0 A0A1L8DPI1 A0A1U8DQI1 A0A0K8W077 K7J676 A0A2Y9LT36 A0A2U4C680 H9JLE7 N6UG27 H0ZH07 A0A091S5W8 A0A2P6L7L4 A0A341CHI8 A0A151WVZ7 A0A1L8DPK9 G1L7L9 A0A154NZ13 A0A2Y9JV52 A0A3L8SAC5 A0A091DS71 G3H1D5 A0A218UVF9 B4HVI6 A0A091IZ45 A0A2J6L2R3 A0A091HZT0 A0A0S3SCE7 A0A195CXH2 Q9W0N8 A0A232EW11 B4H4A7 Q29DW4 A0A226MSY2 F1PSP6 A0A094K4Z6
EC Number
3.4.16.-
Pubmed
26354079
25315136
17994087
17550304
22936249
10731132
+ More
12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30400621 22516182 18362917 19820115 25830018 24495485 15632085 26561354 26483478 12040188 15489334 11447226 16141072 19468303 21183079 20566863 21347285 23371554 20075255 22118469 28004739 23749191 22293439 19121390 23537049 20360741 20010809 30282656 21804562 28401891 26616024 28648823 16341006
12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30400621 22516182 18362917 19820115 25830018 24495485 15632085 26561354 26483478 12040188 15489334 11447226 16141072 19468303 21183079 20566863 21347285 23371554 20075255 22118469 28004739 23749191 22293439 19121390 23537049 20360741 20010809 30282656 21804562 28401891 26616024 28648823 16341006
EMBL
NWSH01000532
PCG75833.1
KQ459896
KPJ19407.1
ODYU01012627
SOQ59044.1
+ More
KQ459551 KPI99925.1 CH954177 EDV58064.1 CM000157 EDW89705.1 CM002910 KMY90425.1 CH480820 EDW54538.1 AE014134 BT072933 AAN10921.1 ACN86081.1 GFDG01001160 JAV17639.1 CH902620 EDV30664.1 CP012523 ALC39891.1 GFDG01001169 JAV17630.1 MK075176 AYV99579.1 BT127423 AEE62385.1 CH916366 EDV96064.1 KQ971312 EEZ98227.1 CH963920 EDW77971.1 CH940654 EDW57620.2 GANO01003086 JAB56785.1 OUUW01000006 SPP82446.1 GFWV01019464 MAA44192.1 GBXI01014206 JAD00086.1 GAMC01017613 JAB88942.1 CH379061 EAL33240.1 CH479228 EDW35379.1 IAAA01001873 LAA03237.1 LSYS01001520 OPJ88568.1 JXUM01050104 KQ561634 KXJ77929.1 CH933809 EDW18057.1 BC004847 BC021399 CH466556 AAH04847.1 AAH21399.1 EDL15861.1 AF330052 AK014680 AK050181 AK153225 AL646096 AL929426 DS235815 EEB17508.1 GBYB01006648 JAG76415.1 CCAG010000500 ADTU01005771 ADTU01005772 ADTU01005773 AKCR02000032 PKK25643.1 CH916368 EDW03650.1 CP012525 ALC43281.1 JXJN01002732 AGBW02012825 OWR44389.1 GEZM01023580 JAV88292.1 GFAH01000131 JAV48258.1 KB742588 EOB06653.1 ADON01023524 AKHW03004113 KYO31021.1 GBBM01000092 JAC35326.1 EEZ98419.1 CH933807 EDW11842.2 EEZ98226.2 CH902618 EDV40191.2 GFDF01005800 JAV08284.1 GDHF01009476 GDHF01007808 JAI42838.1 JAI44506.1 BABH01037195 APGK01036625 KB740941 KB632409 ENN77592.1 ERL95175.1 ABQF01044729 ABQF01044730 KK943506 KFQ52540.1 MWRG01001197 PRD34546.1 KQ982691 KYQ52053.1 GFDF01005799 JAV08285.1 ACTA01083730 ACTA01091730 ACTA01099730 KQ434783 KZC04812.1 QUSF01000033 RLV99264.1 KN121896 KFO34999.1 JH000104 EGV99577.1 MUZQ01000120 OWK57689.1 CH480817 EDW49951.1 KK501252 KFP13632.1 NBSK01005209 PLY81601.1 KL218030 KFP01427.1 AP015039 BAT90485.1 KQ977141 KYN05363.1 AE014296 AY051562 AAF47405.1 AAK92986.1 NNAY01001926 OXU22542.1 CH479208 EDW31222.1 CH379070 EAL30300.2 MCFN01000472 OXB58368.1 AAEX03006560 KL341139 KFZ53007.1
KQ459551 KPI99925.1 CH954177 EDV58064.1 CM000157 EDW89705.1 CM002910 KMY90425.1 CH480820 EDW54538.1 AE014134 BT072933 AAN10921.1 ACN86081.1 GFDG01001160 JAV17639.1 CH902620 EDV30664.1 CP012523 ALC39891.1 GFDG01001169 JAV17630.1 MK075176 AYV99579.1 BT127423 AEE62385.1 CH916366 EDV96064.1 KQ971312 EEZ98227.1 CH963920 EDW77971.1 CH940654 EDW57620.2 GANO01003086 JAB56785.1 OUUW01000006 SPP82446.1 GFWV01019464 MAA44192.1 GBXI01014206 JAD00086.1 GAMC01017613 JAB88942.1 CH379061 EAL33240.1 CH479228 EDW35379.1 IAAA01001873 LAA03237.1 LSYS01001520 OPJ88568.1 JXUM01050104 KQ561634 KXJ77929.1 CH933809 EDW18057.1 BC004847 BC021399 CH466556 AAH04847.1 AAH21399.1 EDL15861.1 AF330052 AK014680 AK050181 AK153225 AL646096 AL929426 DS235815 EEB17508.1 GBYB01006648 JAG76415.1 CCAG010000500 ADTU01005771 ADTU01005772 ADTU01005773 AKCR02000032 PKK25643.1 CH916368 EDW03650.1 CP012525 ALC43281.1 JXJN01002732 AGBW02012825 OWR44389.1 GEZM01023580 JAV88292.1 GFAH01000131 JAV48258.1 KB742588 EOB06653.1 ADON01023524 AKHW03004113 KYO31021.1 GBBM01000092 JAC35326.1 EEZ98419.1 CH933807 EDW11842.2 EEZ98226.2 CH902618 EDV40191.2 GFDF01005800 JAV08284.1 GDHF01009476 GDHF01007808 JAI42838.1 JAI44506.1 BABH01037195 APGK01036625 KB740941 KB632409 ENN77592.1 ERL95175.1 ABQF01044729 ABQF01044730 KK943506 KFQ52540.1 MWRG01001197 PRD34546.1 KQ982691 KYQ52053.1 GFDF01005799 JAV08285.1 ACTA01083730 ACTA01091730 ACTA01099730 KQ434783 KZC04812.1 QUSF01000033 RLV99264.1 KN121896 KFO34999.1 JH000104 EGV99577.1 MUZQ01000120 OWK57689.1 CH480817 EDW49951.1 KK501252 KFP13632.1 NBSK01005209 PLY81601.1 KL218030 KFP01427.1 AP015039 BAT90485.1 KQ977141 KYN05363.1 AE014296 AY051562 AAF47405.1 AAK92986.1 NNAY01001926 OXU22542.1 CH479208 EDW31222.1 CH379070 EAL30300.2 MCFN01000472 OXB58368.1 AAEX03006560 KL341139 KFZ53007.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000192223
UP000095301
UP000008711
+ More
UP000002282 UP000001292 UP000000803 UP000007801 UP000092553 UP000192221 UP000001070 UP000007266 UP000007798 UP000008792 UP000268350 UP000001819 UP000008744 UP000190648 UP000069940 UP000249989 UP000009192 UP000000589 UP000009046 UP000092444 UP000005205 UP000053872 UP000078200 UP000092460 UP000002358 UP000007151 UP000092443 UP000092445 UP000016666 UP000050525 UP000007646 UP000095300 UP000245340 UP000189705 UP000248483 UP000245320 UP000005204 UP000019118 UP000030742 UP000007754 UP000252040 UP000075809 UP000008912 UP000076502 UP000248482 UP000276834 UP000028990 UP000001075 UP000197619 UP000053119 UP000235145 UP000054308 UP000291084 UP000078542 UP000215335 UP000198323 UP000002254
UP000002282 UP000001292 UP000000803 UP000007801 UP000092553 UP000192221 UP000001070 UP000007266 UP000007798 UP000008792 UP000268350 UP000001819 UP000008744 UP000190648 UP000069940 UP000249989 UP000009192 UP000000589 UP000009046 UP000092444 UP000005205 UP000053872 UP000078200 UP000092460 UP000002358 UP000007151 UP000092443 UP000092445 UP000016666 UP000050525 UP000007646 UP000095300 UP000245340 UP000189705 UP000248483 UP000245320 UP000005204 UP000019118 UP000030742 UP000007754 UP000252040 UP000075809 UP000008912 UP000076502 UP000248482 UP000276834 UP000028990 UP000001075 UP000197619 UP000053119 UP000235145 UP000054308 UP000291084 UP000078542 UP000215335 UP000198323 UP000002254
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
A0A2A4JWG7
A0A194RPH1
A0A2H1X335
A0A194Q928
A0A1W4XBY5
A0A1I8M8P6
+ More
B3N626 A0A1W4X145 A0A1I8NA52 B4NXT0 A0A0J9TP36 B4I1Y1 Q8IP31 A0A1L8EGA9 B3ML06 A0A0M4E2M3 A0A1W4VYE8 A0A1L8EFY1 A0A3G5BIH1 J3JW05 B4J2S3 D6W8Z8 B4N036 B4M8U4 U5EQW7 A0A3B0K9X1 A0A293MTR0 A0A0A1WN20 W8AS88 Q29KD6 B4H9E7 A0A1I8M708 A0A2L2Y5G2 A0A1V4KW23 A0A182GA02 B4L0B1 Q99J29 Q920A5 E0VVV2 A0A0C9PYP4 A0A1B0FRI3 A0A158P0S0 A0A2I0M7K2 B4JED0 A0A1A9VMJ8 A0A0M5J3G2 A0A1B0ASD3 K7IP64 A0A212ESD4 A0A1Y1MRG9 A0A1A9Y3F6 A0A1B0AEJ3 A0A1W7RAV1 R0M1S7 U3IBT5 A0A151N2K0 G3T5D9 A0A023GNA5 D6W901 B4KE97 A0A1I8PQA5 D6W8Z9 K7IP63 A0A2U3WA82 A0A1D5PBP3 B3MAE0 A0A1L8DPI1 A0A1U8DQI1 A0A0K8W077 K7J676 A0A2Y9LT36 A0A2U4C680 H9JLE7 N6UG27 H0ZH07 A0A091S5W8 A0A2P6L7L4 A0A341CHI8 A0A151WVZ7 A0A1L8DPK9 G1L7L9 A0A154NZ13 A0A2Y9JV52 A0A3L8SAC5 A0A091DS71 G3H1D5 A0A218UVF9 B4HVI6 A0A091IZ45 A0A2J6L2R3 A0A091HZT0 A0A0S3SCE7 A0A195CXH2 Q9W0N8 A0A232EW11 B4H4A7 Q29DW4 A0A226MSY2 F1PSP6 A0A094K4Z6
B3N626 A0A1W4X145 A0A1I8NA52 B4NXT0 A0A0J9TP36 B4I1Y1 Q8IP31 A0A1L8EGA9 B3ML06 A0A0M4E2M3 A0A1W4VYE8 A0A1L8EFY1 A0A3G5BIH1 J3JW05 B4J2S3 D6W8Z8 B4N036 B4M8U4 U5EQW7 A0A3B0K9X1 A0A293MTR0 A0A0A1WN20 W8AS88 Q29KD6 B4H9E7 A0A1I8M708 A0A2L2Y5G2 A0A1V4KW23 A0A182GA02 B4L0B1 Q99J29 Q920A5 E0VVV2 A0A0C9PYP4 A0A1B0FRI3 A0A158P0S0 A0A2I0M7K2 B4JED0 A0A1A9VMJ8 A0A0M5J3G2 A0A1B0ASD3 K7IP64 A0A212ESD4 A0A1Y1MRG9 A0A1A9Y3F6 A0A1B0AEJ3 A0A1W7RAV1 R0M1S7 U3IBT5 A0A151N2K0 G3T5D9 A0A023GNA5 D6W901 B4KE97 A0A1I8PQA5 D6W8Z9 K7IP63 A0A2U3WA82 A0A1D5PBP3 B3MAE0 A0A1L8DPI1 A0A1U8DQI1 A0A0K8W077 K7J676 A0A2Y9LT36 A0A2U4C680 H9JLE7 N6UG27 H0ZH07 A0A091S5W8 A0A2P6L7L4 A0A341CHI8 A0A151WVZ7 A0A1L8DPK9 G1L7L9 A0A154NZ13 A0A2Y9JV52 A0A3L8SAC5 A0A091DS71 G3H1D5 A0A218UVF9 B4HVI6 A0A091IZ45 A0A2J6L2R3 A0A091HZT0 A0A0S3SCE7 A0A195CXH2 Q9W0N8 A0A232EW11 B4H4A7 Q29DW4 A0A226MSY2 F1PSP6 A0A094K4Z6
PDB
4MWT
E-value=3.77374e-20,
Score=243
Ontologies
GO
PANTHER
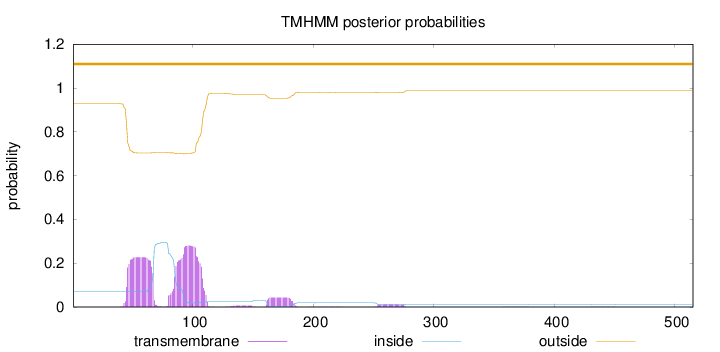
Topology
Subcellular location
Secreted
Length:
515
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
12.42618
Exp number, first 60 AAs:
3.43332
Total prob of N-in:
0.07085
outside
1 - 515
Population Genetic Test Statistics
Pi
249.951326
Theta
195.677907
Tajima's D
0.984469
CLR
0.093268
CSRT
0.652267386630669
Interpretation
Uncertain
