Gene
KWMTBOMO03412 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006376
Annotation
p23-like_protein_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.07
Sequence
CDS
ATGTCTAGCGAAATTTCAGTTCCACCGCCAGTATTATGGGCACAACGAAAGGAAGATGTTTTTCTTACATTTAGTGTGGAAACTAAAGATCCGACGATCAAAATAGAAAAGGATTCAGTGTATTTTAGAGGTGAAGGTGCGCCGGATAATAGACTACATGAGGTAACGATAGCACTGTACGACACTGTTCTTCCGGAAAAGAGTGTTTTTGTAAATAAGGGCAGATGTGTGGAGATGATATTACGGAAGGAAAAAACAGATGGTCCATTCTGGCCGACGCTAACAAAAGACAAGAAAAAGCCGCATTACCTGAAAATTGATTTCAACAAATGGAATGATGAGGATGATGAAGTTGAAGAAAATAATTCATATGATTTCGAGCAAATGCTCCAGAACTTTGGTGGAGCTGGTGGTCCTGGGCAGAAGGCAGAAAAACCTTCGTTTGATGACTTGGAAACTGACTCTGATGATGAGAATTTACCTGATCTGGAATAA
Protein
MSSEISVPPPVLWAQRKEDVFLTFSVETKDPTIKIEKDSVYFRGEGAPDNRLHEVTIALYDTVLPEKSVFVNKGRCVEMILRKEKTDGPFWPTLTKDKKKPHYLKIDFNKWNDEDDEVEENNSYDFEQMLQNFGGAGGPGQKAEKPSFDDLETDSDDENLPDLE
Summary
Uniprot
Q5CCL6
A0A2H1WE07
A0A2A4JVI8
A0A1E1WG54
S4NYB5
A0A0L7L772
+ More
A0A212FBQ4 A0A194RTF6 I4DJJ1 D6WJF6 E2AJL0 A0A0J7NFH2 A0A067QPA7 A0A2A4JX49 A0A026WBJ7 A0A3L8DTH9 A0A1B6HUY6 A0A2J7QVH9 A0A2J7QVH0 A0A1B6CKZ7 A0A1B6M6L6 A0A1B6CPB7 R4UMY2 V5H153 A0A1Y1N096 A0A158P357 E2BX56 A0A088ARS8 A0A170Z2A9 A0A2H1WYE5 A0A224XM34 A0A069DNN7 A0A195F2R5 A0A2A3E3L5 V9IL14 A0A151WFH7 A0A1E1W0Z9 A0A0P4VSE5 R4G851 A0A0T6B1L5 A0A0V0G5E8 A0A0L7RDC4 A0A0N0U2R2 S4PWL3 T1JH99 A0A2P2HW88 H9JDZ2 A0A195BD20 A0A182WDI8 A0A087T9F5 U5ESS5 A0A3Q0JGP4 A0A195DDR1 A0A151IGE1 A0A0C9RNA8 A0A182QV73 A0A2R5LH61 A0A182KL42 A0A147BA41 A0A182JKP0 A0A2M3Z136 A0A182V9K8 A0A182X5M6 Q7QHE6 A0A182HWC3 T1E8M2 A0A182RTD0 A0A182LUY1 A0A0K8TPP8 U4UER5 A0A1Z5L6D4 A0A182KBU8 M9VTU4 A0A182UKV5 A0A2M4A8B0 W5J5U0 A0A293LGR1 A0A2M4C1H4 A0A182FKK9 N6UF92 R4WNB2 A0A182NZM4 A0A182N2B5 A0A182YB23 A0A1W7R9X9 E7D1W6 A0A182SNF6 V3ZZ19 A0A023G824 A0A2L2Y2D7 A0A336MGZ7
A0A212FBQ4 A0A194RTF6 I4DJJ1 D6WJF6 E2AJL0 A0A0J7NFH2 A0A067QPA7 A0A2A4JX49 A0A026WBJ7 A0A3L8DTH9 A0A1B6HUY6 A0A2J7QVH9 A0A2J7QVH0 A0A1B6CKZ7 A0A1B6M6L6 A0A1B6CPB7 R4UMY2 V5H153 A0A1Y1N096 A0A158P357 E2BX56 A0A088ARS8 A0A170Z2A9 A0A2H1WYE5 A0A224XM34 A0A069DNN7 A0A195F2R5 A0A2A3E3L5 V9IL14 A0A151WFH7 A0A1E1W0Z9 A0A0P4VSE5 R4G851 A0A0T6B1L5 A0A0V0G5E8 A0A0L7RDC4 A0A0N0U2R2 S4PWL3 T1JH99 A0A2P2HW88 H9JDZ2 A0A195BD20 A0A182WDI8 A0A087T9F5 U5ESS5 A0A3Q0JGP4 A0A195DDR1 A0A151IGE1 A0A0C9RNA8 A0A182QV73 A0A2R5LH61 A0A182KL42 A0A147BA41 A0A182JKP0 A0A2M3Z136 A0A182V9K8 A0A182X5M6 Q7QHE6 A0A182HWC3 T1E8M2 A0A182RTD0 A0A182LUY1 A0A0K8TPP8 U4UER5 A0A1Z5L6D4 A0A182KBU8 M9VTU4 A0A182UKV5 A0A2M4A8B0 W5J5U0 A0A293LGR1 A0A2M4C1H4 A0A182FKK9 N6UF92 R4WNB2 A0A182NZM4 A0A182N2B5 A0A182YB23 A0A1W7R9X9 E7D1W6 A0A182SNF6 V3ZZ19 A0A023G824 A0A2L2Y2D7 A0A336MGZ7
Pubmed
EMBL
BABH01009253
AB206399
BAD90845.1
ODYU01007909
SOQ51102.1
NWSH01000532
+ More
PCG75829.1 GDQN01005082 JAT85972.1 GAIX01013865 JAA78695.1 JTDY01002482 KOB71328.1 AGBW02009294 OWR51148.1 KQ459896 KPJ19401.1 AK401459 KQ459551 BAM18081.1 KPI99932.1 KQ971342 EFA03889.1 GL440027 EFN66405.1 LBMM01005677 KMQ91295.1 KK853097 KDR11450.1 NWSH01000412 PCG76571.1 KK107293 EZA53333.1 QOIP01000005 RLU23068.1 GECU01029217 JAS78489.1 NEVH01010475 PNF32583.1 PNF32582.1 GEDC01023159 JAS14139.1 GEBQ01019645 GEBQ01008419 JAT20332.1 JAT31558.1 GEDC01021976 JAS15322.1 KC571976 AGM32475.1 GALX01001898 JAB66568.1 GEZM01016200 JAV91341.1 ADTU01001303 GL451202 EFN79775.1 GEMB01002688 JAS00507.1 ODYU01012016 SOQ58093.1 GFTR01002880 JAW13546.1 GBGD01003206 JAC85683.1 KQ981855 KYN34753.1 KZ288427 PBC25882.1 JR050040 AEY61161.1 KQ983219 KYQ46535.1 GDQN01010386 GDQN01005966 JAT80668.1 JAT85088.1 GDKW01001208 JAI55387.1 ACPB03000870 GAHY01001582 JAA75928.1 LJIG01016180 KRT81376.1 GECL01003549 JAP02575.1 KQ414614 KOC68824.1 KQ438551 KOX67174.1 GAIX01006588 JAA85972.1 JH432223 IACF01000019 LAB65836.1 BABH01002561 KQ976522 KYM82085.1 KK114126 KFM61744.1 GANO01003099 JAB56772.1 KQ980989 KYN10564.1 KQ977726 KYN00294.1 GBYB01008566 JAG78333.1 AXCN02001729 GGLE01004662 MBY08788.1 GEIB01000100 JAR87659.1 GGFM01001496 MBW22247.1 AAAB01008816 EAA05132.4 APCN01001484 GAMD01002633 JAA98957.1 AXCM01001359 GDAI01001500 JAI16103.1 KB632288 ERL91527.1 GFJQ02004115 JAW02855.1 KC261579 AGJ83338.1 GGFK01003715 MBW37036.1 ADMH02002090 ETN59356.1 GFWV01003192 MAA27922.1 GGFJ01009983 MBW59124.1 APGK01037418 KB740948 ENN77327.1 AK417101 BAN20316.1 GFAH01000430 JAV47959.1 HQ006070 ADV40360.1 KB203115 ESO86251.1 GBBM01006363 JAC29055.1 IAAA01000434 IAAA01000436 IAAA01000437 LAA02336.1 UFQS01000939 UFQT01000939 SSX07859.1 SSX28093.1
PCG75829.1 GDQN01005082 JAT85972.1 GAIX01013865 JAA78695.1 JTDY01002482 KOB71328.1 AGBW02009294 OWR51148.1 KQ459896 KPJ19401.1 AK401459 KQ459551 BAM18081.1 KPI99932.1 KQ971342 EFA03889.1 GL440027 EFN66405.1 LBMM01005677 KMQ91295.1 KK853097 KDR11450.1 NWSH01000412 PCG76571.1 KK107293 EZA53333.1 QOIP01000005 RLU23068.1 GECU01029217 JAS78489.1 NEVH01010475 PNF32583.1 PNF32582.1 GEDC01023159 JAS14139.1 GEBQ01019645 GEBQ01008419 JAT20332.1 JAT31558.1 GEDC01021976 JAS15322.1 KC571976 AGM32475.1 GALX01001898 JAB66568.1 GEZM01016200 JAV91341.1 ADTU01001303 GL451202 EFN79775.1 GEMB01002688 JAS00507.1 ODYU01012016 SOQ58093.1 GFTR01002880 JAW13546.1 GBGD01003206 JAC85683.1 KQ981855 KYN34753.1 KZ288427 PBC25882.1 JR050040 AEY61161.1 KQ983219 KYQ46535.1 GDQN01010386 GDQN01005966 JAT80668.1 JAT85088.1 GDKW01001208 JAI55387.1 ACPB03000870 GAHY01001582 JAA75928.1 LJIG01016180 KRT81376.1 GECL01003549 JAP02575.1 KQ414614 KOC68824.1 KQ438551 KOX67174.1 GAIX01006588 JAA85972.1 JH432223 IACF01000019 LAB65836.1 BABH01002561 KQ976522 KYM82085.1 KK114126 KFM61744.1 GANO01003099 JAB56772.1 KQ980989 KYN10564.1 KQ977726 KYN00294.1 GBYB01008566 JAG78333.1 AXCN02001729 GGLE01004662 MBY08788.1 GEIB01000100 JAR87659.1 GGFM01001496 MBW22247.1 AAAB01008816 EAA05132.4 APCN01001484 GAMD01002633 JAA98957.1 AXCM01001359 GDAI01001500 JAI16103.1 KB632288 ERL91527.1 GFJQ02004115 JAW02855.1 KC261579 AGJ83338.1 GGFK01003715 MBW37036.1 ADMH02002090 ETN59356.1 GFWV01003192 MAA27922.1 GGFJ01009983 MBW59124.1 APGK01037418 KB740948 ENN77327.1 AK417101 BAN20316.1 GFAH01000430 JAV47959.1 HQ006070 ADV40360.1 KB203115 ESO86251.1 GBBM01006363 JAC29055.1 IAAA01000434 IAAA01000436 IAAA01000437 LAA02336.1 UFQS01000939 UFQT01000939 SSX07859.1 SSX28093.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053240
UP000053268
+ More
UP000007266 UP000000311 UP000036403 UP000027135 UP000053097 UP000279307 UP000235965 UP000005205 UP000008237 UP000005203 UP000078541 UP000242457 UP000075809 UP000015103 UP000053825 UP000053105 UP000078540 UP000075920 UP000054359 UP000079169 UP000078492 UP000078542 UP000075886 UP000075882 UP000075880 UP000075903 UP000076407 UP000007062 UP000075840 UP000075900 UP000075883 UP000030742 UP000075881 UP000075902 UP000000673 UP000069272 UP000019118 UP000075885 UP000075884 UP000076408 UP000075901 UP000030746
UP000007266 UP000000311 UP000036403 UP000027135 UP000053097 UP000279307 UP000235965 UP000005205 UP000008237 UP000005203 UP000078541 UP000242457 UP000075809 UP000015103 UP000053825 UP000053105 UP000078540 UP000075920 UP000054359 UP000079169 UP000078492 UP000078542 UP000075886 UP000075882 UP000075880 UP000075903 UP000076407 UP000007062 UP000075840 UP000075900 UP000075883 UP000030742 UP000075881 UP000075902 UP000000673 UP000069272 UP000019118 UP000075885 UP000075884 UP000076408 UP000075901 UP000030746
Pfam
PF04969 CS
SUPFAM
SSF49764
SSF49764
Gene 3D
ProteinModelPortal
Q5CCL6
A0A2H1WE07
A0A2A4JVI8
A0A1E1WG54
S4NYB5
A0A0L7L772
+ More
A0A212FBQ4 A0A194RTF6 I4DJJ1 D6WJF6 E2AJL0 A0A0J7NFH2 A0A067QPA7 A0A2A4JX49 A0A026WBJ7 A0A3L8DTH9 A0A1B6HUY6 A0A2J7QVH9 A0A2J7QVH0 A0A1B6CKZ7 A0A1B6M6L6 A0A1B6CPB7 R4UMY2 V5H153 A0A1Y1N096 A0A158P357 E2BX56 A0A088ARS8 A0A170Z2A9 A0A2H1WYE5 A0A224XM34 A0A069DNN7 A0A195F2R5 A0A2A3E3L5 V9IL14 A0A151WFH7 A0A1E1W0Z9 A0A0P4VSE5 R4G851 A0A0T6B1L5 A0A0V0G5E8 A0A0L7RDC4 A0A0N0U2R2 S4PWL3 T1JH99 A0A2P2HW88 H9JDZ2 A0A195BD20 A0A182WDI8 A0A087T9F5 U5ESS5 A0A3Q0JGP4 A0A195DDR1 A0A151IGE1 A0A0C9RNA8 A0A182QV73 A0A2R5LH61 A0A182KL42 A0A147BA41 A0A182JKP0 A0A2M3Z136 A0A182V9K8 A0A182X5M6 Q7QHE6 A0A182HWC3 T1E8M2 A0A182RTD0 A0A182LUY1 A0A0K8TPP8 U4UER5 A0A1Z5L6D4 A0A182KBU8 M9VTU4 A0A182UKV5 A0A2M4A8B0 W5J5U0 A0A293LGR1 A0A2M4C1H4 A0A182FKK9 N6UF92 R4WNB2 A0A182NZM4 A0A182N2B5 A0A182YB23 A0A1W7R9X9 E7D1W6 A0A182SNF6 V3ZZ19 A0A023G824 A0A2L2Y2D7 A0A336MGZ7
A0A212FBQ4 A0A194RTF6 I4DJJ1 D6WJF6 E2AJL0 A0A0J7NFH2 A0A067QPA7 A0A2A4JX49 A0A026WBJ7 A0A3L8DTH9 A0A1B6HUY6 A0A2J7QVH9 A0A2J7QVH0 A0A1B6CKZ7 A0A1B6M6L6 A0A1B6CPB7 R4UMY2 V5H153 A0A1Y1N096 A0A158P357 E2BX56 A0A088ARS8 A0A170Z2A9 A0A2H1WYE5 A0A224XM34 A0A069DNN7 A0A195F2R5 A0A2A3E3L5 V9IL14 A0A151WFH7 A0A1E1W0Z9 A0A0P4VSE5 R4G851 A0A0T6B1L5 A0A0V0G5E8 A0A0L7RDC4 A0A0N0U2R2 S4PWL3 T1JH99 A0A2P2HW88 H9JDZ2 A0A195BD20 A0A182WDI8 A0A087T9F5 U5ESS5 A0A3Q0JGP4 A0A195DDR1 A0A151IGE1 A0A0C9RNA8 A0A182QV73 A0A2R5LH61 A0A182KL42 A0A147BA41 A0A182JKP0 A0A2M3Z136 A0A182V9K8 A0A182X5M6 Q7QHE6 A0A182HWC3 T1E8M2 A0A182RTD0 A0A182LUY1 A0A0K8TPP8 U4UER5 A0A1Z5L6D4 A0A182KBU8 M9VTU4 A0A182UKV5 A0A2M4A8B0 W5J5U0 A0A293LGR1 A0A2M4C1H4 A0A182FKK9 N6UF92 R4WNB2 A0A182NZM4 A0A182N2B5 A0A182YB23 A0A1W7R9X9 E7D1W6 A0A182SNF6 V3ZZ19 A0A023G824 A0A2L2Y2D7 A0A336MGZ7
PDB
1EJF
E-value=2.48492e-09,
Score=143
Ontologies
PATHWAY
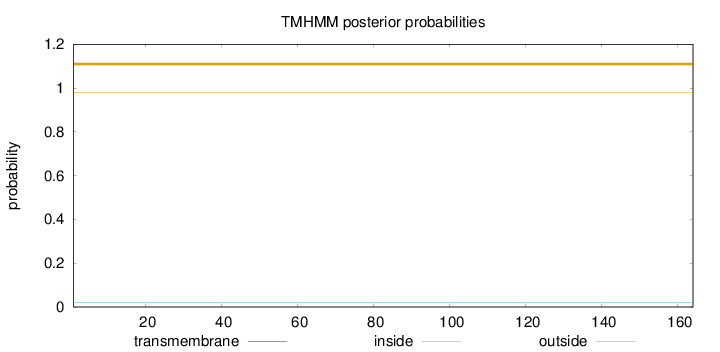
Topology
Length:
164
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00021
Exp number, first 60 AAs:
9e-05
Total prob of N-in:
0.01971
outside
1 - 164
Population Genetic Test Statistics
Pi
230.156089
Theta
208.864773
Tajima's D
0.34888
CLR
0
CSRT
0.468626568671566
Interpretation
Uncertain
