Gene
KWMTBOMO03411
Pre Gene Modal
BGIBMGA004264
Annotation
Retrovirus-related_Pol_polyprotein_from_transposon_17.6_[Exaiptasia_pallida]
Location in the cell
Nuclear Reliability : 3.106
Sequence
CDS
ATGACTGAAACACCCACAACAAACATGGTGTCAGGTGTGGGGTTCCTGCCACTTTTACCCAATATGGATTTCAAAGGTAACCTTGCCGAAAATTTCAAAATGTATCGTCAAAGGTTGGAGATATATTTGTCTGCTAATGGCCTTGATGACGCCAAAGACAAAAGAAAAATATCAATTCTGCTGAATTTTATTGGCGAAAACGGAATCAAAATCTACAATTCTTTCAACTTAAACGCATCTGAATCACATACTTTTGAACACGTGCTCAAAAAGTTTGAGGCGTACTTTGAACCAAAGAAAAGTCTAACTATAGCAAGGAACGAGTTTTTTACTGTTACACAAGATGAAGACAGGTTGGAGGATTACTTTCAAAGAGTAATAAATCTCGGTATTCAATGCGAACTTGGAGATCTCAGAGAAGGGTTGACAGTGGGAAAGATAATTTCAGGGTTAGACTCCAAATATAATAATCTCAAAACACGATTAATGTCGGAAGACGATAGTAAATTAACATTAGATTATGTTATGAAATACTTACTGAGTGCCGAAACCTCTAGAAAATATATAGAAGAAAAAACAAATTCCAAAGACAGCGATGAATCCTCCATCCTATATGTTAATAAAAAATCAATAAGGAACTGTAGAAACTGTGGCACATCTCATAATATAAATAAATGCCCAGCATACGGCAAGGAGTGTCTGAAGTGCAAGAAACTTAACCATTTTAGTAAAGTATGCAGATCAGCCAAGAAACCAAATTTTTATCAAAAGCAAGTCAATGTCCTTGAGGATCCAGATAAGAACAAAGAGGATGAAGCTCAAGGAGATTTCTTTGTTGGTTATGCAGGCAATGAAATAATTCAAGATTTTACAGTCAATAACATCAAAATTCCCATCAAAATGGACACTGGAGCTGCATGCAACGTGATTGGGAGCGAATGA
Protein
MTETPTTNMVSGVGFLPLLPNMDFKGNLAENFKMYRQRLEIYLSANGLDDAKDKRKISILLNFIGENGIKIYNSFNLNASESHTFEHVLKKFEAYFEPKKSLTIARNEFFTVTQDEDRLEDYFQRVINLGIQCELGDLREGLTVGKIISGLDSKYNNLKTRLMSEDDSKLTLDYVMKYLLSAETSRKYIEEKTNSKDSDESSILYVNKKSIRNCRNCGTSHNINKCPAYGKECLKCKKLNHFSKVCRSAKKPNFYQKQVNVLEDPDKNKEDEAQGDFFVGYAGNEIIQDFTVNNIKIPIKMDTGAACNVIGSE
Summary
Uniprot
A0A2G8L9H5
A0A3P8Q6C6
A0A3B3C9X5
A0A3P9HT22
A0A3P9M8I3
A0A3B3HLX7
+ More
A0A2G8KKG5 X1WTY2 A0A1A8UGA0 A0A087U9K1 A0A3B1JKR1 A0A2S2N8I3 A0A3P8S414 A0A212FHD4 A0A2S2NL10 A0A1A8IVJ0 A0A1S4E819 A0A1Y1MUX5 A0A1A8D4P4 A0A146SN99 A0A1Y1M671 A0A146S5H9 A0A1S3DM59 A0A3B3I1S4 A0A3Q0J4U3 A0A3P9HXP1 A0A1Y1MCJ7 A0A069DY08 A0A069DZS8 A0A1Y1NN34 L7LWW6 A0A0A9Z4Y4 A0A1A8JBP0 A0A2A4K615 A0A1Y1LR12 J9M4C2 A0A3P8QAD1 A0A2G8LMB1 A0A146LC45 A0A146TAZ6 I3KX76 A0A3Q0J653 A0A1I8NEP4 A0A075EIL5 A0A1I8NEN3 A0A1I8NEQ0 A0A1I8NEP7 A0A1Y1NHV7 A0A2S2NI46 I3KYE2 A0A2G8KBJ0 X1XQZ7 T1I2W9 A0A023EY30 A0A3P9AYU2 T1IA98 A0A3P8RKT7 A0A2G8KB94 A0A1S3JEC7 A0A3P8PD24 A0A3Q0JLN5 A0A1S3DS68 A0A3P8RHF8 J9LNB8 A0A151IWV0 A0A3Q0J548 A0A1S3ICH8 A0A3Q0JFZ1 A0A2G8KHQ8 J9LDY4 A0A1B8XWD8 J9JQ26 A0A1Y1LNL5 A0A1Y1JV53 A0A087UXZ8 A0A1Y1KPK9 T1HVY1 A0A1Y1LZ99 A0A2G8KFU3 A0A1L8H7D0 A0A1Y1K454 A0A1S3HTD1 A0A3Q0JKH1 A0A1A8B469 J9LYD2 A0A2G8L310 A0A2G8KVE9 A0A151X058 A0A224Z9B6 A0A1S3DNR0 J9K926 A0A1Y1LB52 A0A1S3DM37 A0A1S3DH71 A0A146PYY4 A0A151JNU9 A0A1Y1NAD7
A0A2G8KKG5 X1WTY2 A0A1A8UGA0 A0A087U9K1 A0A3B1JKR1 A0A2S2N8I3 A0A3P8S414 A0A212FHD4 A0A2S2NL10 A0A1A8IVJ0 A0A1S4E819 A0A1Y1MUX5 A0A1A8D4P4 A0A146SN99 A0A1Y1M671 A0A146S5H9 A0A1S3DM59 A0A3B3I1S4 A0A3Q0J4U3 A0A3P9HXP1 A0A1Y1MCJ7 A0A069DY08 A0A069DZS8 A0A1Y1NN34 L7LWW6 A0A0A9Z4Y4 A0A1A8JBP0 A0A2A4K615 A0A1Y1LR12 J9M4C2 A0A3P8QAD1 A0A2G8LMB1 A0A146LC45 A0A146TAZ6 I3KX76 A0A3Q0J653 A0A1I8NEP4 A0A075EIL5 A0A1I8NEN3 A0A1I8NEQ0 A0A1I8NEP7 A0A1Y1NHV7 A0A2S2NI46 I3KYE2 A0A2G8KBJ0 X1XQZ7 T1I2W9 A0A023EY30 A0A3P9AYU2 T1IA98 A0A3P8RKT7 A0A2G8KB94 A0A1S3JEC7 A0A3P8PD24 A0A3Q0JLN5 A0A1S3DS68 A0A3P8RHF8 J9LNB8 A0A151IWV0 A0A3Q0J548 A0A1S3ICH8 A0A3Q0JFZ1 A0A2G8KHQ8 J9LDY4 A0A1B8XWD8 J9JQ26 A0A1Y1LNL5 A0A1Y1JV53 A0A087UXZ8 A0A1Y1KPK9 T1HVY1 A0A1Y1LZ99 A0A2G8KFU3 A0A1L8H7D0 A0A1Y1K454 A0A1S3HTD1 A0A3Q0JKH1 A0A1A8B469 J9LYD2 A0A2G8L310 A0A2G8KVE9 A0A151X058 A0A224Z9B6 A0A1S3DNR0 J9K926 A0A1Y1LB52 A0A1S3DM37 A0A1S3DH71 A0A146PYY4 A0A151JNU9 A0A1Y1NAD7
Pubmed
EMBL
MRZV01000161
PIK56894.1
MRZV01000519
PIK48493.1
ABLF02013930
ABLF02013934
+ More
HADY01023773 HAEJ01006887 SBS47344.1 KK118858 KFM74040.1 GGMR01000868 MBY13487.1 AGBW02008504 OWR53150.1 GGMR01005013 MBY17632.1 HAED01014930 SBR01375.1 GEZM01020000 JAV89472.1 HADZ01022257 SBP86198.1 GCES01104253 JAQ82069.1 GEZM01041944 JAV80090.1 GCES01110503 JAQ75819.1 GEZM01035137 JAV83383.1 GBGD01000119 JAC88770.1 GBGD01000115 JAC88774.1 GEZM01001974 JAV97427.1 GACK01009641 JAA55393.1 GBHO01004090 JAG39514.1 HAED01020456 SBR07020.1 NWSH01000088 PCG79705.1 GEZM01050758 JAV75411.1 ABLF02038957 MRZV01000033 PIK61397.1 GDHC01013957 JAQ04672.1 GCES01096219 JAQ90103.1 AERX01076149 KF319019 AIE48224.1 GEZM01001973 JAV97431.1 GGMR01004179 MBY16798.1 AERX01070080 MRZV01000718 PIK45343.1 ABLF02041865 ACPB03002101 ACPB03002102 ACPB03002103 GBBI01004745 JAC13967.1 ACPB03019169 MRZV01000723 PIK45255.1 ABLF02008456 KQ980844 KYN12280.1 MRZV01000576 PIK47528.1 ABLF02010707 KV461054 OCA14957.1 ABLF02019772 GEZM01050997 GEZM01050995 JAV75219.1 GEZM01102055 GEZM01102054 GEZM01102053 GEZM01102051 JAV52011.1 KK122208 KFM82237.1 GEZM01079607 GEZM01079598 JAV62391.1 ACPB03023527 GEZM01043682 JAV78912.1 MRZV01000618 PIK46835.1 CM004469 OCT91956.1 GEZM01097791 JAV54106.1 HADY01023564 SBP62049.1 ABLF02041541 MRZV01000245 PIK54530.1 MRZV01000347 PIK51998.1 KQ982635 KYQ53341.1 GFPF01012437 MAA23583.1 ABLF02008619 GEZM01060849 JAV70854.1 GCES01137294 JAQ49028.1 KQ978837 KYN27997.1 GEZM01011167 JAV93615.1
HADY01023773 HAEJ01006887 SBS47344.1 KK118858 KFM74040.1 GGMR01000868 MBY13487.1 AGBW02008504 OWR53150.1 GGMR01005013 MBY17632.1 HAED01014930 SBR01375.1 GEZM01020000 JAV89472.1 HADZ01022257 SBP86198.1 GCES01104253 JAQ82069.1 GEZM01041944 JAV80090.1 GCES01110503 JAQ75819.1 GEZM01035137 JAV83383.1 GBGD01000119 JAC88770.1 GBGD01000115 JAC88774.1 GEZM01001974 JAV97427.1 GACK01009641 JAA55393.1 GBHO01004090 JAG39514.1 HAED01020456 SBR07020.1 NWSH01000088 PCG79705.1 GEZM01050758 JAV75411.1 ABLF02038957 MRZV01000033 PIK61397.1 GDHC01013957 JAQ04672.1 GCES01096219 JAQ90103.1 AERX01076149 KF319019 AIE48224.1 GEZM01001973 JAV97431.1 GGMR01004179 MBY16798.1 AERX01070080 MRZV01000718 PIK45343.1 ABLF02041865 ACPB03002101 ACPB03002102 ACPB03002103 GBBI01004745 JAC13967.1 ACPB03019169 MRZV01000723 PIK45255.1 ABLF02008456 KQ980844 KYN12280.1 MRZV01000576 PIK47528.1 ABLF02010707 KV461054 OCA14957.1 ABLF02019772 GEZM01050997 GEZM01050995 JAV75219.1 GEZM01102055 GEZM01102054 GEZM01102053 GEZM01102051 JAV52011.1 KK122208 KFM82237.1 GEZM01079607 GEZM01079598 JAV62391.1 ACPB03023527 GEZM01043682 JAV78912.1 MRZV01000618 PIK46835.1 CM004469 OCT91956.1 GEZM01097791 JAV54106.1 HADY01023564 SBP62049.1 ABLF02041541 MRZV01000245 PIK54530.1 MRZV01000347 PIK51998.1 KQ982635 KYQ53341.1 GFPF01012437 MAA23583.1 ABLF02008619 GEZM01060849 JAV70854.1 GCES01137294 JAQ49028.1 KQ978837 KYN27997.1 GEZM01011167 JAV93615.1
Proteomes
Pfam
Interpro
IPR021109
Peptidase_aspartic_dom_sf
+ More
IPR001584 Integrase_cat-core
IPR041577 RT_RNaseH_2
IPR001878 Znf_CCHC
IPR012337 RNaseH-like_sf
IPR000477 RT_dom
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR001995 Peptidase_A2_cat
IPR001969 Aspartic_peptidase_AS
IPR001781 Znf_LIM
IPR003128 Villin_headpiece
IPR036886 Villin_headpiece_dom_sf
IPR036875 Znf_CCHC_sf
IPR041373 RT_RNaseH
IPR001584 Integrase_cat-core
IPR041577 RT_RNaseH_2
IPR001878 Znf_CCHC
IPR012337 RNaseH-like_sf
IPR000477 RT_dom
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR001995 Peptidase_A2_cat
IPR001969 Aspartic_peptidase_AS
IPR001781 Znf_LIM
IPR003128 Villin_headpiece
IPR036886 Villin_headpiece_dom_sf
IPR036875 Znf_CCHC_sf
IPR041373 RT_RNaseH
Gene 3D
ProteinModelPortal
A0A2G8L9H5
A0A3P8Q6C6
A0A3B3C9X5
A0A3P9HT22
A0A3P9M8I3
A0A3B3HLX7
+ More
A0A2G8KKG5 X1WTY2 A0A1A8UGA0 A0A087U9K1 A0A3B1JKR1 A0A2S2N8I3 A0A3P8S414 A0A212FHD4 A0A2S2NL10 A0A1A8IVJ0 A0A1S4E819 A0A1Y1MUX5 A0A1A8D4P4 A0A146SN99 A0A1Y1M671 A0A146S5H9 A0A1S3DM59 A0A3B3I1S4 A0A3Q0J4U3 A0A3P9HXP1 A0A1Y1MCJ7 A0A069DY08 A0A069DZS8 A0A1Y1NN34 L7LWW6 A0A0A9Z4Y4 A0A1A8JBP0 A0A2A4K615 A0A1Y1LR12 J9M4C2 A0A3P8QAD1 A0A2G8LMB1 A0A146LC45 A0A146TAZ6 I3KX76 A0A3Q0J653 A0A1I8NEP4 A0A075EIL5 A0A1I8NEN3 A0A1I8NEQ0 A0A1I8NEP7 A0A1Y1NHV7 A0A2S2NI46 I3KYE2 A0A2G8KBJ0 X1XQZ7 T1I2W9 A0A023EY30 A0A3P9AYU2 T1IA98 A0A3P8RKT7 A0A2G8KB94 A0A1S3JEC7 A0A3P8PD24 A0A3Q0JLN5 A0A1S3DS68 A0A3P8RHF8 J9LNB8 A0A151IWV0 A0A3Q0J548 A0A1S3ICH8 A0A3Q0JFZ1 A0A2G8KHQ8 J9LDY4 A0A1B8XWD8 J9JQ26 A0A1Y1LNL5 A0A1Y1JV53 A0A087UXZ8 A0A1Y1KPK9 T1HVY1 A0A1Y1LZ99 A0A2G8KFU3 A0A1L8H7D0 A0A1Y1K454 A0A1S3HTD1 A0A3Q0JKH1 A0A1A8B469 J9LYD2 A0A2G8L310 A0A2G8KVE9 A0A151X058 A0A224Z9B6 A0A1S3DNR0 J9K926 A0A1Y1LB52 A0A1S3DM37 A0A1S3DH71 A0A146PYY4 A0A151JNU9 A0A1Y1NAD7
A0A2G8KKG5 X1WTY2 A0A1A8UGA0 A0A087U9K1 A0A3B1JKR1 A0A2S2N8I3 A0A3P8S414 A0A212FHD4 A0A2S2NL10 A0A1A8IVJ0 A0A1S4E819 A0A1Y1MUX5 A0A1A8D4P4 A0A146SN99 A0A1Y1M671 A0A146S5H9 A0A1S3DM59 A0A3B3I1S4 A0A3Q0J4U3 A0A3P9HXP1 A0A1Y1MCJ7 A0A069DY08 A0A069DZS8 A0A1Y1NN34 L7LWW6 A0A0A9Z4Y4 A0A1A8JBP0 A0A2A4K615 A0A1Y1LR12 J9M4C2 A0A3P8QAD1 A0A2G8LMB1 A0A146LC45 A0A146TAZ6 I3KX76 A0A3Q0J653 A0A1I8NEP4 A0A075EIL5 A0A1I8NEN3 A0A1I8NEQ0 A0A1I8NEP7 A0A1Y1NHV7 A0A2S2NI46 I3KYE2 A0A2G8KBJ0 X1XQZ7 T1I2W9 A0A023EY30 A0A3P9AYU2 T1IA98 A0A3P8RKT7 A0A2G8KB94 A0A1S3JEC7 A0A3P8PD24 A0A3Q0JLN5 A0A1S3DS68 A0A3P8RHF8 J9LNB8 A0A151IWV0 A0A3Q0J548 A0A1S3ICH8 A0A3Q0JFZ1 A0A2G8KHQ8 J9LDY4 A0A1B8XWD8 J9JQ26 A0A1Y1LNL5 A0A1Y1JV53 A0A087UXZ8 A0A1Y1KPK9 T1HVY1 A0A1Y1LZ99 A0A2G8KFU3 A0A1L8H7D0 A0A1Y1K454 A0A1S3HTD1 A0A3Q0JKH1 A0A1A8B469 J9LYD2 A0A2G8L310 A0A2G8KVE9 A0A151X058 A0A224Z9B6 A0A1S3DNR0 J9K926 A0A1Y1LB52 A0A1S3DM37 A0A1S3DH71 A0A146PYY4 A0A151JNU9 A0A1Y1NAD7
PDB
2BL6
E-value=0.0371663,
Score=85
Ontologies
PATHWAY
GO
Topology
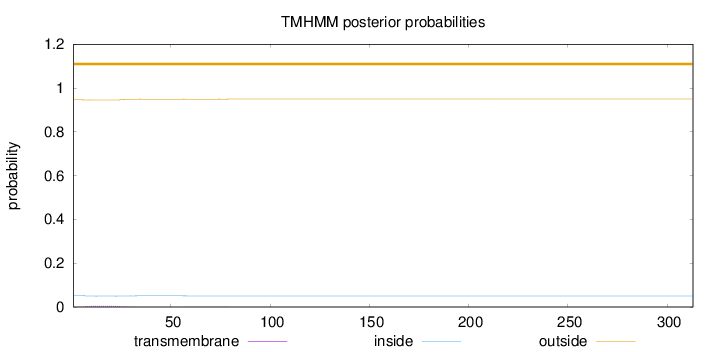
Length:
313
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.104
Exp number, first 60 AAs:
0.0965599999999999
Total prob of N-in:
0.05387
outside
1 - 313
Population Genetic Test Statistics
Pi
230.308268
Theta
209.347594
Tajima's D
0.315212
CLR
0
CSRT
0.455227238638068
Interpretation
Uncertain
