Gene
KWMTBOMO03403
Pre Gene Modal
BGIBMGA006498
Annotation
PREDICTED:_protein-lysine_methyltransferase_METTL21D-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 0.769
Sequence
CDS
ATGTGCCAATATAAATCTGATTTTCTGAGTGGATTAAGAGTTCTAGAACTTGGAGCTGGACTTGGAGTTGTTGGTCTAACCGCTGCGACACTTGGAGCTCAAGTTACAATTACAGACTTACCAGAAGCTCTACCGTTGATACGATTGAATATCAACGAAAATAAGTCCAAGATTGGTAGTATGGGTGGCTACGCAACTGCAGAATCACTTATTTGGGGAGATCAGTATTCTGAAATATTCAATGAAGAATATGATTTAATAGTTTTAGCTGATTGTATTTATTATGAAGAAGCAATAGAGCAATTGCTAATAACACTTCAGCTTTTAAGTAGAAGTCTGAAAAAAAAGCCGGTGATTTACTTAACACAAGAAATTAGAGATTCTGAGATACAAAAAAGGCTTTGGAATTTATTTTATGAAAAACTGTCTGAATCATTTTCCATTGAGAAAATACCAGAAGAAGAGCAGCATAGTAACTATAGAAGTGAAGATATAGTTTTATTAAGATTGGAAAAAAAGTAA
Protein
MCQYKSDFLSGLRVLELGAGLGVVGLTAATLGAQVTITDLPEALPLIRLNINENKSKIGSMGGYATAESLIWGDQYSEIFNEEYDLIVLADCIYYEEAIEQLLITLQLLSRSLKKKPVIYLTQEIRDSEIQKRLWNLFYEKLSESFSIEKIPEEEQHSNYRSEDIVLLRLEKK
Summary
Uniprot
A0A1E1WH20
A0A212FBM5
A0A2H1X3G8
A0A194Q2Z1
A0A0L7KYM9
A0A2J7QHH1
+ More
K7J153 A0A232FHT5 J9JKT2 A0A2S2PUZ8 A0A2H8TLK1 A0A1B6JF98 V4BBG2 A0A1B6GTJ3 A0A067RA54 A0A1B6DCC2 A0A1B6L7E1 A0A1B6C5F0 A0A2R7WUQ6 E0VA00 A0A210QLG8 A0A023F945 R4FKI5 A0A069DQX5 A0A2I4BLY7 A0A0V0G9V4 E9H2Z2 A0A1A8SFS5 A0A1A8QVI0 A0A1A8QRQ3 C3Y1R5 A0A1I7YN04 A0A1A8F1K9 A0A1A8I4C1 A0A1A8V3D5 A0A3P8UEX6 K1PWK2 A0A2L2YD69 H2VAQ8 A0A3P8UIA5 A0A0P5V851 A0A0P5K781 A0A0P5PQT9 A0A2B4SUD3 A0A0P5SMI9 A0A3P9D089 V9LFK7 A0A0P5XU09 A0A2L2YAX9 A0A3B4H5M0 A0A3P8QXH7 A0A3B5A1C9 A0A0P5V0I5 A0A0P5RVC0 A0A3B3CUG6 W5UML2 A0A0P5VEN2 A0A0P5WA97 A0A0P5B4P5 A0A0C9R183 A0A0P5DYI6 A0A0N8D7Z2 A0A0P6A7D7 A0A0P5K2B7 A0A0P5V9N1 I3KW94 A0A0N8CFF5 R7VBY5 A0A0P6JSE4 A0A023G4J0 A0A0C9S001 G3NYY1 A0A091III1 G3TQS6 R7VT97 A0A2P6KRL0 A0A293N2K6 A0A1V4L069 A0A2I0LSH7 A0A147B4Y7 A0A099Z367 A0A3P9KF46 A0A087FXP6 A0A147BMT6 A0A0P5VVQ5 A0A369SRJ0 A0A1U7UID9 G1RN89 A0A2Y9QJD0 A0A091EY70 A0A2Y9DAR2 A0A091ITP4 A0A2U3ZPN5 A0A3P9HFG3 A0A2U3WH69 A0A2Y9EN03 A0A1W5B254 A0A093GIL1
K7J153 A0A232FHT5 J9JKT2 A0A2S2PUZ8 A0A2H8TLK1 A0A1B6JF98 V4BBG2 A0A1B6GTJ3 A0A067RA54 A0A1B6DCC2 A0A1B6L7E1 A0A1B6C5F0 A0A2R7WUQ6 E0VA00 A0A210QLG8 A0A023F945 R4FKI5 A0A069DQX5 A0A2I4BLY7 A0A0V0G9V4 E9H2Z2 A0A1A8SFS5 A0A1A8QVI0 A0A1A8QRQ3 C3Y1R5 A0A1I7YN04 A0A1A8F1K9 A0A1A8I4C1 A0A1A8V3D5 A0A3P8UEX6 K1PWK2 A0A2L2YD69 H2VAQ8 A0A3P8UIA5 A0A0P5V851 A0A0P5K781 A0A0P5PQT9 A0A2B4SUD3 A0A0P5SMI9 A0A3P9D089 V9LFK7 A0A0P5XU09 A0A2L2YAX9 A0A3B4H5M0 A0A3P8QXH7 A0A3B5A1C9 A0A0P5V0I5 A0A0P5RVC0 A0A3B3CUG6 W5UML2 A0A0P5VEN2 A0A0P5WA97 A0A0P5B4P5 A0A0C9R183 A0A0P5DYI6 A0A0N8D7Z2 A0A0P6A7D7 A0A0P5K2B7 A0A0P5V9N1 I3KW94 A0A0N8CFF5 R7VBY5 A0A0P6JSE4 A0A023G4J0 A0A0C9S001 G3NYY1 A0A091III1 G3TQS6 R7VT97 A0A2P6KRL0 A0A293N2K6 A0A1V4L069 A0A2I0LSH7 A0A147B4Y7 A0A099Z367 A0A3P9KF46 A0A087FXP6 A0A147BMT6 A0A0P5VVQ5 A0A369SRJ0 A0A1U7UID9 G1RN89 A0A2Y9QJD0 A0A091EY70 A0A2Y9DAR2 A0A091ITP4 A0A2U3ZPN5 A0A3P9HFG3 A0A2U3WH69 A0A2Y9EN03 A0A1W5B254 A0A093GIL1
Pubmed
EMBL
GDQN01004796
JAT86258.1
AGBW02009294
OWR51144.1
ODYU01013148
SOQ59839.1
+ More
KQ459551 KPI99927.1 JTDY01004356 KOB68255.1 NEVH01013976 PNF28031.1 NNAY01000180 OXU30242.1 ABLF02026377 GGMR01020535 MBY33154.1 GFXV01002363 MBW14168.1 GECU01009882 JAS97824.1 KB200027 ESP03382.1 GECZ01004005 JAS65764.1 KK852785 KDR16562.1 GEDC01013955 JAS23343.1 GEBQ01020402 JAT19575.1 GEDC01028833 JAS08465.1 KK855386 PTY22305.1 DS235002 EEB10206.1 NEDP02003088 OWF49511.1 GBBI01000706 JAC18006.1 ACPB03016356 GAHY01001833 JAA75677.1 GBGD01002837 JAC86052.1 GECL01001162 JAP04962.1 GL732587 EFX73928.1 HAEI01014309 SBS16778.1 HAEH01013382 SBR97263.1 HAEF01018829 HAEG01014006 SBR96455.1 GG666480 EEN65805.1 HAEB01006475 SBQ53002.1 HAED01005676 SBQ91706.1 HADY01022914 HAEJ01013311 SBS53768.1 JH816124 EKC20720.1 IAAA01016717 LAA05300.1 GDIP01103541 JAM00174.1 GDIQ01193770 JAK57955.1 GDIQ01124681 JAL27045.1 LSMT01000026 PFX32188.1 GDIQ01093704 JAL58022.1 JW878841 AFP11358.1 GDIP01067315 JAM36400.1 IAAA01016714 IAAA01016715 IAAA01016716 LAA05293.1 GDIP01106235 JAL97479.1 GDIQ01095757 JAL55969.1 JT415080 AHH40912.1 GDIP01116160 JAL87554.1 GDIP01088968 JAM14747.1 GDIP01190200 JAJ33202.1 GBZX01002025 JAG90715.1 GDIP01167246 JAJ56156.1 GDIP01059911 GDIQ01048234 LRGB01001831 JAM43804.1 JAN46503.1 KZS10353.1 GDIP01033712 JAM70003.1 GDIQ01195672 JAK56053.1 GDIP01103540 JAM00175.1 AERX01030271 GDIP01137007 JAL66707.1 AMQN01004378 KB293367 ELU16077.1 GDIQ01010361 JAN84376.1 GBBM01006272 JAC29146.1 GBYB01013721 JAG83488.1 KL218647 KFP07313.1 KB375459 EMC87258.1 MWRG01006470 PRD28942.1 GFWV01020887 MAA45615.1 LSYS01000634 OPJ90035.1 AKCR02000112 PKK20382.1 GCES01000477 JAR85846.1 KL889586 KGL76894.1 KL989625 KFK22398.1 GEGO01003612 JAR91792.1 GDIP01094515 JAM09200.1 NOWV01000001 RDD47793.1 ADFV01040044 KK719186 KFO61946.1 KK500855 KFP10913.1 KL215798 KFV66629.1
KQ459551 KPI99927.1 JTDY01004356 KOB68255.1 NEVH01013976 PNF28031.1 NNAY01000180 OXU30242.1 ABLF02026377 GGMR01020535 MBY33154.1 GFXV01002363 MBW14168.1 GECU01009882 JAS97824.1 KB200027 ESP03382.1 GECZ01004005 JAS65764.1 KK852785 KDR16562.1 GEDC01013955 JAS23343.1 GEBQ01020402 JAT19575.1 GEDC01028833 JAS08465.1 KK855386 PTY22305.1 DS235002 EEB10206.1 NEDP02003088 OWF49511.1 GBBI01000706 JAC18006.1 ACPB03016356 GAHY01001833 JAA75677.1 GBGD01002837 JAC86052.1 GECL01001162 JAP04962.1 GL732587 EFX73928.1 HAEI01014309 SBS16778.1 HAEH01013382 SBR97263.1 HAEF01018829 HAEG01014006 SBR96455.1 GG666480 EEN65805.1 HAEB01006475 SBQ53002.1 HAED01005676 SBQ91706.1 HADY01022914 HAEJ01013311 SBS53768.1 JH816124 EKC20720.1 IAAA01016717 LAA05300.1 GDIP01103541 JAM00174.1 GDIQ01193770 JAK57955.1 GDIQ01124681 JAL27045.1 LSMT01000026 PFX32188.1 GDIQ01093704 JAL58022.1 JW878841 AFP11358.1 GDIP01067315 JAM36400.1 IAAA01016714 IAAA01016715 IAAA01016716 LAA05293.1 GDIP01106235 JAL97479.1 GDIQ01095757 JAL55969.1 JT415080 AHH40912.1 GDIP01116160 JAL87554.1 GDIP01088968 JAM14747.1 GDIP01190200 JAJ33202.1 GBZX01002025 JAG90715.1 GDIP01167246 JAJ56156.1 GDIP01059911 GDIQ01048234 LRGB01001831 JAM43804.1 JAN46503.1 KZS10353.1 GDIP01033712 JAM70003.1 GDIQ01195672 JAK56053.1 GDIP01103540 JAM00175.1 AERX01030271 GDIP01137007 JAL66707.1 AMQN01004378 KB293367 ELU16077.1 GDIQ01010361 JAN84376.1 GBBM01006272 JAC29146.1 GBYB01013721 JAG83488.1 KL218647 KFP07313.1 KB375459 EMC87258.1 MWRG01006470 PRD28942.1 GFWV01020887 MAA45615.1 LSYS01000634 OPJ90035.1 AKCR02000112 PKK20382.1 GCES01000477 JAR85846.1 KL889586 KGL76894.1 KL989625 KFK22398.1 GEGO01003612 JAR91792.1 GDIP01094515 JAM09200.1 NOWV01000001 RDD47793.1 ADFV01040044 KK719186 KFO61946.1 KK500855 KFP10913.1 KL215798 KFV66629.1
Proteomes
UP000007151
UP000053268
UP000037510
UP000235965
UP000002358
UP000215335
+ More
UP000007819 UP000030746 UP000027135 UP000009046 UP000242188 UP000015103 UP000192220 UP000000305 UP000001554 UP000095287 UP000265080 UP000005408 UP000005226 UP000265120 UP000225706 UP000265160 UP000261460 UP000265100 UP000261400 UP000261560 UP000221080 UP000076858 UP000005207 UP000014760 UP000007635 UP000054308 UP000007646 UP000190648 UP000053872 UP000053641 UP000265180 UP000253843 UP000189704 UP000001073 UP000248480 UP000052976 UP000053119 UP000245340 UP000265200 UP000248484 UP000192224 UP000053875
UP000007819 UP000030746 UP000027135 UP000009046 UP000242188 UP000015103 UP000192220 UP000000305 UP000001554 UP000095287 UP000265080 UP000005408 UP000005226 UP000265120 UP000225706 UP000265160 UP000261460 UP000265100 UP000261400 UP000261560 UP000221080 UP000076858 UP000005207 UP000014760 UP000007635 UP000054308 UP000007646 UP000190648 UP000053872 UP000053641 UP000265180 UP000253843 UP000189704 UP000001073 UP000248480 UP000052976 UP000053119 UP000245340 UP000265200 UP000248484 UP000192224 UP000053875
Pfam
PF10294 Methyltransf_16
SUPFAM
SSF53335
SSF53335
ProteinModelPortal
A0A1E1WH20
A0A212FBM5
A0A2H1X3G8
A0A194Q2Z1
A0A0L7KYM9
A0A2J7QHH1
+ More
K7J153 A0A232FHT5 J9JKT2 A0A2S2PUZ8 A0A2H8TLK1 A0A1B6JF98 V4BBG2 A0A1B6GTJ3 A0A067RA54 A0A1B6DCC2 A0A1B6L7E1 A0A1B6C5F0 A0A2R7WUQ6 E0VA00 A0A210QLG8 A0A023F945 R4FKI5 A0A069DQX5 A0A2I4BLY7 A0A0V0G9V4 E9H2Z2 A0A1A8SFS5 A0A1A8QVI0 A0A1A8QRQ3 C3Y1R5 A0A1I7YN04 A0A1A8F1K9 A0A1A8I4C1 A0A1A8V3D5 A0A3P8UEX6 K1PWK2 A0A2L2YD69 H2VAQ8 A0A3P8UIA5 A0A0P5V851 A0A0P5K781 A0A0P5PQT9 A0A2B4SUD3 A0A0P5SMI9 A0A3P9D089 V9LFK7 A0A0P5XU09 A0A2L2YAX9 A0A3B4H5M0 A0A3P8QXH7 A0A3B5A1C9 A0A0P5V0I5 A0A0P5RVC0 A0A3B3CUG6 W5UML2 A0A0P5VEN2 A0A0P5WA97 A0A0P5B4P5 A0A0C9R183 A0A0P5DYI6 A0A0N8D7Z2 A0A0P6A7D7 A0A0P5K2B7 A0A0P5V9N1 I3KW94 A0A0N8CFF5 R7VBY5 A0A0P6JSE4 A0A023G4J0 A0A0C9S001 G3NYY1 A0A091III1 G3TQS6 R7VT97 A0A2P6KRL0 A0A293N2K6 A0A1V4L069 A0A2I0LSH7 A0A147B4Y7 A0A099Z367 A0A3P9KF46 A0A087FXP6 A0A147BMT6 A0A0P5VVQ5 A0A369SRJ0 A0A1U7UID9 G1RN89 A0A2Y9QJD0 A0A091EY70 A0A2Y9DAR2 A0A091ITP4 A0A2U3ZPN5 A0A3P9HFG3 A0A2U3WH69 A0A2Y9EN03 A0A1W5B254 A0A093GIL1
K7J153 A0A232FHT5 J9JKT2 A0A2S2PUZ8 A0A2H8TLK1 A0A1B6JF98 V4BBG2 A0A1B6GTJ3 A0A067RA54 A0A1B6DCC2 A0A1B6L7E1 A0A1B6C5F0 A0A2R7WUQ6 E0VA00 A0A210QLG8 A0A023F945 R4FKI5 A0A069DQX5 A0A2I4BLY7 A0A0V0G9V4 E9H2Z2 A0A1A8SFS5 A0A1A8QVI0 A0A1A8QRQ3 C3Y1R5 A0A1I7YN04 A0A1A8F1K9 A0A1A8I4C1 A0A1A8V3D5 A0A3P8UEX6 K1PWK2 A0A2L2YD69 H2VAQ8 A0A3P8UIA5 A0A0P5V851 A0A0P5K781 A0A0P5PQT9 A0A2B4SUD3 A0A0P5SMI9 A0A3P9D089 V9LFK7 A0A0P5XU09 A0A2L2YAX9 A0A3B4H5M0 A0A3P8QXH7 A0A3B5A1C9 A0A0P5V0I5 A0A0P5RVC0 A0A3B3CUG6 W5UML2 A0A0P5VEN2 A0A0P5WA97 A0A0P5B4P5 A0A0C9R183 A0A0P5DYI6 A0A0N8D7Z2 A0A0P6A7D7 A0A0P5K2B7 A0A0P5V9N1 I3KW94 A0A0N8CFF5 R7VBY5 A0A0P6JSE4 A0A023G4J0 A0A0C9S001 G3NYY1 A0A091III1 G3TQS6 R7VT97 A0A2P6KRL0 A0A293N2K6 A0A1V4L069 A0A2I0LSH7 A0A147B4Y7 A0A099Z367 A0A3P9KF46 A0A087FXP6 A0A147BMT6 A0A0P5VVQ5 A0A369SRJ0 A0A1U7UID9 G1RN89 A0A2Y9QJD0 A0A091EY70 A0A2Y9DAR2 A0A091ITP4 A0A2U3ZPN5 A0A3P9HFG3 A0A2U3WH69 A0A2Y9EN03 A0A1W5B254 A0A093GIL1
PDB
4LG1
E-value=2.23352e-12,
Score=170
Ontologies
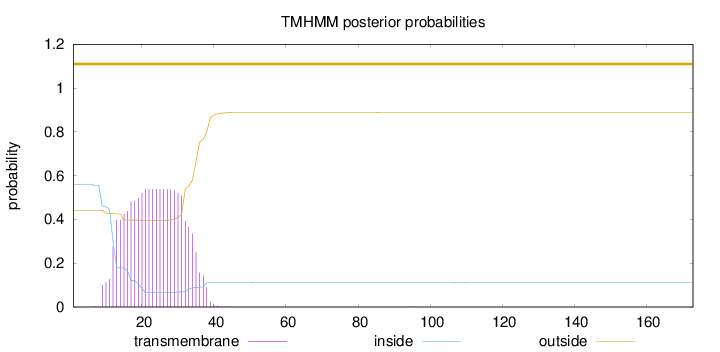
Topology
Length:
173
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
11.92425
Exp number, first 60 AAs:
11.91052
Total prob of N-in:
0.55848
POSSIBLE N-term signal
sequence
outside
1 - 173
Population Genetic Test Statistics
Pi
217.600702
Theta
203.814283
Tajima's D
0.21293
CLR
21.922493
CSRT
0.431528423578821
Interpretation
Uncertain
