Gene
KWMTBOMO03399
Pre Gene Modal
BGIBMGA006496
Annotation
PREDICTED:_TAF5-like_RNA_polymerase_II_p300/CBP-associated_factor-associated_factor_65_kDa_subunit_5L_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 1.872
Sequence
CDS
ATGTCATTAATAAAGAAGACAAGAAACGATGCAATTAAAGCTGCGGTGGCTTCTTATCTAGAAAGGCGAAACTATCCCGATACTGAATATTACGGTAACAATAACCCAAATTGCCAGAGTGCTGAGCAAATGGCAGTTTCCACTATTGTACAGTCTGAAGCAAGTCGTGCAAATTCTATCTTGTTTTCATGTATCAACAATGATTCCTCTCAATACGATATACAATACACCAGGTTAGTCAACTTTATAAAAGAAATAAGAATCGAGAAACTAAAAAATGAGCTTCTGGGATTGTTGAGCCCTTTGCTATGTCATCTTTATCTAGAGATGCTACGTGGGGAACACGGAGGTGCTGCACAAGTGTTTTTGAAACGACATTCAGCAACATTACCACAGAAAGATTTATCTTTCCATCAACCAATTGATGGGAATTTACCCTCTGCATTGTATAGGCCAAATAGTCTAGAGCAACTGTTTAATTCACTTCAAAATGGTACAATAGATAATGAAACCCCCGAAAAAGATTACATGAATCAAATACTTGATGATATTGGATCCATCTACACTTTACAGGACATTGAAACTAGACCATCCATTGCTGCATTCAGGTCATGTAAATATGATATTTACTTATCACAAGATGCTTTAAATATGTTGAAAGCATATTTAGCCAAACATGGACATGTATTATTAATACAAGTATTGCAAACATGGTTTCATATTGATATAAATAGTGATAATAATAAAAAAAATTCTGAAGACGAAGAAGAAATGGAGTCTGATATGACAGTTAATAGAAATTTTTCTGAAGAACGAGTCATAGAAGCAAACGACCTGTTTTCTAAATGTAATGGCCATGCTGAATATCAAACTGTTGATAAGGAGTTGCGTGATCTCCAAGATGCAATCAAAGGAGTTAGAGACTCAACGGCTCCATTAAAATTGTACAAAATATCAGCACCGGATACGCATTTAATATGTGCAAAAACAGATCCTAGCTGTAACATGCTGTGCGGTGGCTTCATTAATTCTGAAGTCAGAATCTGGGATCTTGGCCAAAACAATATGCAAAAAAGGATCAATCATAACATATCAAAAATTGAATTAGCTTGCAACATGCCACCAGAATCTTCAAACGATCATGATACATTACAAATTGGCGCCGGATTGCCCCTGAGAGGCCACTCAGGCCCAGTACAAGCTGTGACTATTATGCCCAGAGAAGAATTAATACTCTCAGCATCACACGATAACACAATGAGAGCTTGGAGACTGTCTGATTATTCATGTGCTGCAATTTACAGAGGACACAATTATCCTATATGGTGTATGGACATATCAAGAAGTGGCCTATTTATAGTAACGGGATCTCATGACAGAACAGCTAAGCTTTGGTCTTTGAATAGAACGTTCCCAGTGAGAGTGTTTGTCGGTCACATGTCTGATGTCACTTGTGTGAAGTTCCACCCGAACGAGTGTTACATAGCGTCTGGCGGCGCGGACCGCAGCGTGCGGCTGTGGAGCGTGTGCGACGGCCGGCTCGCACGCGTCCTGTGCGCACACCGCGCGCCGGTGCGGGCGCTCGCCTTCGCACCCAGCGGAGCCTACCTAGCCACTGCCGGAGACGATAAAAAAATTAAAGTGTGGGACTTAGCAGCAGGAACTTGCGTGCAAGAGTACAAAGGACACCATGGTAAAATAATGTCACTGGATTGGTCTTCGGTCGGAAAACCCAGCCTAACTAACAGAGTAATGGCAGAACCTAATGAATTATCAACGGAAAACTCTCTACTCTGTTCAGCAGGAATGGATGGAATTGTGAAAGTTTTTCATGATTGTCAGACTAAAAATAAGCCAACCTCGAGTCAAGATATTTTAACATCAACTTATAATACGAAATGCACCTATTTGGTGGATGTACAAGTGCATCCGGAATGGGTTGTTGCGATTGGAACCAAGCAATGA
Protein
MSLIKKTRNDAIKAAVASYLERRNYPDTEYYGNNNPNCQSAEQMAVSTIVQSEASRANSILFSCINNDSSQYDIQYTRLVNFIKEIRIEKLKNELLGLLSPLLCHLYLEMLRGEHGGAAQVFLKRHSATLPQKDLSFHQPIDGNLPSALYRPNSLEQLFNSLQNGTIDNETPEKDYMNQILDDIGSIYTLQDIETRPSIAAFRSCKYDIYLSQDALNMLKAYLAKHGHVLLIQVLQTWFHIDINSDNNKKNSEDEEEMESDMTVNRNFSEERVIEANDLFSKCNGHAEYQTVDKELRDLQDAIKGVRDSTAPLKLYKISAPDTHLICAKTDPSCNMLCGGFINSEVRIWDLGQNNMQKRINHNISKIELACNMPPESSNDHDTLQIGAGLPLRGHSGPVQAVTIMPREELILSASHDNTMRAWRLSDYSCAAIYRGHNYPIWCMDISRSGLFIVTGSHDRTAKLWSLNRTFPVRVFVGHMSDVTCVKFHPNECYIASGGADRSVRLWSVCDGRLARVLCAHRAPVRALAFAPSGAYLATAGDDKKIKVWDLAAGTCVQEYKGHHGKIMSLDWSSVGKPSLTNRVMAEPNELSTENSLLCSAGMDGIVKVFHDCQTKNKPTSSQDILTSTYNTKCTYLVDVQVHPEWVVAIGTKQ
Summary
Uniprot
A0A0N0PCM3
A0A2H1VL01
A0A2A4JUX2
A0A2A4JVX0
A0A212FBN0
H9JAF1
+ More
A0A194Q320 A0A2J7PNX2 A0A067QRZ3 E0VPQ2 A0A1B6M6T8 A0A232FFU3 A0A1B6DIH6 A0A0C9R6A3 A0A3L8D867 A0A026WIS8 K7IYA5 A0A158NN42 A0A151J657 F4X1W0 A0A151WKT2 A0A0L7R830 E2BW42 E2A2U2 A0A195BB43 E9IDU0 A0A195FAC4 A0A195C2X4 A0A2J7PNU9 A0A2I4CRM3 A0A310SAF4 A0A3P8S4Y5 F1QIA7 A0A3B4ZYP7 A0A3P8VW69 A0A1A8SDI2 A0A1A8M4G6 A0A1A7YQB2 A0A1A8AV83 A0A1A8FS18 A0A1A8I9P4 Q6PGV3 A0A091UNW6 A0A1A8CZL3 H2SF11 Q6DE31 A0A3B4T610 A0A3P9IFX2 A0A3B4XWZ3 A0A3B3S904 A0A1S3AQG8 Q32NS1 A0A131Z1E1 A0A224Z1S4 A0A3P9LE43 U3I667 A0A3B3HTB1 A0A091L8S4 A0A0F8ASG4 A0A0F8CDY2 A0A2I0MUY5 T1J6X2 V8NTP9 A0A3P8Y1I9 A0A1V4JKN8 A0A0N8K2Y7 A0A091GRT6 A0A093SD50 A0A1S3PUM2 A0A1W4ZRP3 A0A091E759 A0A3B5QDC7 A0A087R6V8 A0A093C1C4 A0A1U7RML9 A0A093GW99 A0A3B4H5R0 A0A3P9BAP0 A0A093IHX6 A0A2I0TZE4 A0A093THY2 A0A087VMR2 A0A091JIJ6 A0A0A0AW15 A0A094K9L4 A0A091G7B1 A0A093PEU5 A0A094KPM5 A0A091PNY6 A0A091SDY0 A0A093I049 A0A218VAS9 A0A099YUS7 M7BA75 A0A3P8Q3L4 A0A091MCG8 U3JL60
A0A194Q320 A0A2J7PNX2 A0A067QRZ3 E0VPQ2 A0A1B6M6T8 A0A232FFU3 A0A1B6DIH6 A0A0C9R6A3 A0A3L8D867 A0A026WIS8 K7IYA5 A0A158NN42 A0A151J657 F4X1W0 A0A151WKT2 A0A0L7R830 E2BW42 E2A2U2 A0A195BB43 E9IDU0 A0A195FAC4 A0A195C2X4 A0A2J7PNU9 A0A2I4CRM3 A0A310SAF4 A0A3P8S4Y5 F1QIA7 A0A3B4ZYP7 A0A3P8VW69 A0A1A8SDI2 A0A1A8M4G6 A0A1A7YQB2 A0A1A8AV83 A0A1A8FS18 A0A1A8I9P4 Q6PGV3 A0A091UNW6 A0A1A8CZL3 H2SF11 Q6DE31 A0A3B4T610 A0A3P9IFX2 A0A3B4XWZ3 A0A3B3S904 A0A1S3AQG8 Q32NS1 A0A131Z1E1 A0A224Z1S4 A0A3P9LE43 U3I667 A0A3B3HTB1 A0A091L8S4 A0A0F8ASG4 A0A0F8CDY2 A0A2I0MUY5 T1J6X2 V8NTP9 A0A3P8Y1I9 A0A1V4JKN8 A0A0N8K2Y7 A0A091GRT6 A0A093SD50 A0A1S3PUM2 A0A1W4ZRP3 A0A091E759 A0A3B5QDC7 A0A087R6V8 A0A093C1C4 A0A1U7RML9 A0A093GW99 A0A3B4H5R0 A0A3P9BAP0 A0A093IHX6 A0A2I0TZE4 A0A093THY2 A0A087VMR2 A0A091JIJ6 A0A0A0AW15 A0A094K9L4 A0A091G7B1 A0A093PEU5 A0A094KPM5 A0A091PNY6 A0A091SDY0 A0A093I049 A0A218VAS9 A0A099YUS7 M7BA75 A0A3P8Q3L4 A0A091MCG8 U3JL60
Pubmed
EMBL
KQ460544
KPJ14101.1
ODYU01003130
SOQ41503.1
NWSH01000532
PCG75837.1
+ More
PCG75838.1 AGBW02009294 OWR51140.1 BABH01009245 BABH01009246 KQ459551 KPI99936.1 NEVH01023284 PNF18016.1 KK853094 KDR11498.1 DS235370 EEB15358.1 GEBQ01008331 JAT31646.1 NNAY01000306 OXU29328.1 GEDC01011795 JAS25503.1 GBYB01003525 JAG73292.1 QOIP01000011 RLU16687.1 KK107182 EZA55947.1 ADTU01021015 KQ979935 KYN18592.1 GL888553 EGI59569.1 KQ982993 KYQ48493.1 KQ414637 KOC66988.1 GL451091 EFN80074.1 GL436214 EFN72249.1 KQ976537 KYM81440.1 GL762535 EFZ21256.1 KQ981693 KYN37575.1 KQ978344 KYM94945.1 PNF18017.1 KQ766607 OAD53621.1 CU467834 HAEH01014752 HAEI01013136 SBS15605.1 HAEF01011099 SBR51663.1 HADW01005278 HADX01010136 SBP32368.1 HADY01019891 HAEJ01004564 SBP58376.1 HAEB01015034 HAEC01001729 SBQ61561.1 HAED01007586 HAEE01013842 SBQ93798.1 BC056820 AAH56820.1 KL409753 KFQ91615.1 HADZ01020120 SBP84061.1 BC077313 CM004474 AAH77313.1 OCT80007.1 BC108505 CM004475 AAI08506.1 OCT77964.1 GEDV01003945 JAP84612.1 GFPF01012621 MAA23767.1 ADON01077260 KK743240 KFP39391.1 KQ041256 KKF28987.1 KQ042248 KKF17080.1 AKCR02000002 PKK33492.1 JH431896 AZIM01001885 ETE65440.1 LSYS01006902 OPJ72756.1 JARO02000303 KPP79049.1 KL509185 KFO85879.1 KL671001 KFW80699.1 KK717998 KFO53923.1 KL226149 KFM09212.1 KL237365 KFV09760.1 KL205735 KFV73666.1 KK594523 KFW02325.1 KZ506558 PKU39149.1 KL439954 KFW93929.1 KL499056 KFO13904.1 KK502085 KFP20402.1 KL873332 KGL98156.1 KL340286 KFZ52461.1 KL447787 KFO77059.1 KL225298 KFW70737.1 KL264298 KFZ61168.1 KK676138 KFQ09018.1 KK469313 KFQ56558.1 KK401265 KFV60078.1 MUZQ01000016 OWK63137.1 KL885573 KGL73106.1 KB594214 EMP25132.1 KK823589 KFP71585.1 AGTO01021885
PCG75838.1 AGBW02009294 OWR51140.1 BABH01009245 BABH01009246 KQ459551 KPI99936.1 NEVH01023284 PNF18016.1 KK853094 KDR11498.1 DS235370 EEB15358.1 GEBQ01008331 JAT31646.1 NNAY01000306 OXU29328.1 GEDC01011795 JAS25503.1 GBYB01003525 JAG73292.1 QOIP01000011 RLU16687.1 KK107182 EZA55947.1 ADTU01021015 KQ979935 KYN18592.1 GL888553 EGI59569.1 KQ982993 KYQ48493.1 KQ414637 KOC66988.1 GL451091 EFN80074.1 GL436214 EFN72249.1 KQ976537 KYM81440.1 GL762535 EFZ21256.1 KQ981693 KYN37575.1 KQ978344 KYM94945.1 PNF18017.1 KQ766607 OAD53621.1 CU467834 HAEH01014752 HAEI01013136 SBS15605.1 HAEF01011099 SBR51663.1 HADW01005278 HADX01010136 SBP32368.1 HADY01019891 HAEJ01004564 SBP58376.1 HAEB01015034 HAEC01001729 SBQ61561.1 HAED01007586 HAEE01013842 SBQ93798.1 BC056820 AAH56820.1 KL409753 KFQ91615.1 HADZ01020120 SBP84061.1 BC077313 CM004474 AAH77313.1 OCT80007.1 BC108505 CM004475 AAI08506.1 OCT77964.1 GEDV01003945 JAP84612.1 GFPF01012621 MAA23767.1 ADON01077260 KK743240 KFP39391.1 KQ041256 KKF28987.1 KQ042248 KKF17080.1 AKCR02000002 PKK33492.1 JH431896 AZIM01001885 ETE65440.1 LSYS01006902 OPJ72756.1 JARO02000303 KPP79049.1 KL509185 KFO85879.1 KL671001 KFW80699.1 KK717998 KFO53923.1 KL226149 KFM09212.1 KL237365 KFV09760.1 KL205735 KFV73666.1 KK594523 KFW02325.1 KZ506558 PKU39149.1 KL439954 KFW93929.1 KL499056 KFO13904.1 KK502085 KFP20402.1 KL873332 KGL98156.1 KL340286 KFZ52461.1 KL447787 KFO77059.1 KL225298 KFW70737.1 KL264298 KFZ61168.1 KK676138 KFQ09018.1 KK469313 KFQ56558.1 KK401265 KFV60078.1 MUZQ01000016 OWK63137.1 KL885573 KGL73106.1 KB594214 EMP25132.1 KK823589 KFP71585.1 AGTO01021885
Proteomes
UP000053240
UP000218220
UP000007151
UP000005204
UP000053268
UP000235965
+ More
UP000027135 UP000009046 UP000215335 UP000279307 UP000053097 UP000002358 UP000005205 UP000078492 UP000007755 UP000075809 UP000053825 UP000008237 UP000000311 UP000078540 UP000078541 UP000078542 UP000192220 UP000265080 UP000000437 UP000261400 UP000265120 UP000053283 UP000005226 UP000186698 UP000261420 UP000265200 UP000261360 UP000261540 UP000079721 UP000265180 UP000016666 UP000001038 UP000053872 UP000265140 UP000190648 UP000034805 UP000053258 UP000087266 UP000192224 UP000052976 UP000002852 UP000053286 UP000189705 UP000053584 UP000261460 UP000265160 UP000053119 UP000053858 UP000053760 UP000054081 UP000197619 UP000053641 UP000031443 UP000265100 UP000016665
UP000027135 UP000009046 UP000215335 UP000279307 UP000053097 UP000002358 UP000005205 UP000078492 UP000007755 UP000075809 UP000053825 UP000008237 UP000000311 UP000078540 UP000078541 UP000078542 UP000192220 UP000265080 UP000000437 UP000261400 UP000265120 UP000053283 UP000005226 UP000186698 UP000261420 UP000265200 UP000261360 UP000261540 UP000079721 UP000265180 UP000016666 UP000001038 UP000053872 UP000265140 UP000190648 UP000034805 UP000053258 UP000087266 UP000192224 UP000052976 UP000002852 UP000053286 UP000189705 UP000053584 UP000261460 UP000265160 UP000053119 UP000053858 UP000053760 UP000054081 UP000197619 UP000053641 UP000031443 UP000265100 UP000016665
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A0N0PCM3
A0A2H1VL01
A0A2A4JUX2
A0A2A4JVX0
A0A212FBN0
H9JAF1
+ More
A0A194Q320 A0A2J7PNX2 A0A067QRZ3 E0VPQ2 A0A1B6M6T8 A0A232FFU3 A0A1B6DIH6 A0A0C9R6A3 A0A3L8D867 A0A026WIS8 K7IYA5 A0A158NN42 A0A151J657 F4X1W0 A0A151WKT2 A0A0L7R830 E2BW42 E2A2U2 A0A195BB43 E9IDU0 A0A195FAC4 A0A195C2X4 A0A2J7PNU9 A0A2I4CRM3 A0A310SAF4 A0A3P8S4Y5 F1QIA7 A0A3B4ZYP7 A0A3P8VW69 A0A1A8SDI2 A0A1A8M4G6 A0A1A7YQB2 A0A1A8AV83 A0A1A8FS18 A0A1A8I9P4 Q6PGV3 A0A091UNW6 A0A1A8CZL3 H2SF11 Q6DE31 A0A3B4T610 A0A3P9IFX2 A0A3B4XWZ3 A0A3B3S904 A0A1S3AQG8 Q32NS1 A0A131Z1E1 A0A224Z1S4 A0A3P9LE43 U3I667 A0A3B3HTB1 A0A091L8S4 A0A0F8ASG4 A0A0F8CDY2 A0A2I0MUY5 T1J6X2 V8NTP9 A0A3P8Y1I9 A0A1V4JKN8 A0A0N8K2Y7 A0A091GRT6 A0A093SD50 A0A1S3PUM2 A0A1W4ZRP3 A0A091E759 A0A3B5QDC7 A0A087R6V8 A0A093C1C4 A0A1U7RML9 A0A093GW99 A0A3B4H5R0 A0A3P9BAP0 A0A093IHX6 A0A2I0TZE4 A0A093THY2 A0A087VMR2 A0A091JIJ6 A0A0A0AW15 A0A094K9L4 A0A091G7B1 A0A093PEU5 A0A094KPM5 A0A091PNY6 A0A091SDY0 A0A093I049 A0A218VAS9 A0A099YUS7 M7BA75 A0A3P8Q3L4 A0A091MCG8 U3JL60
A0A194Q320 A0A2J7PNX2 A0A067QRZ3 E0VPQ2 A0A1B6M6T8 A0A232FFU3 A0A1B6DIH6 A0A0C9R6A3 A0A3L8D867 A0A026WIS8 K7IYA5 A0A158NN42 A0A151J657 F4X1W0 A0A151WKT2 A0A0L7R830 E2BW42 E2A2U2 A0A195BB43 E9IDU0 A0A195FAC4 A0A195C2X4 A0A2J7PNU9 A0A2I4CRM3 A0A310SAF4 A0A3P8S4Y5 F1QIA7 A0A3B4ZYP7 A0A3P8VW69 A0A1A8SDI2 A0A1A8M4G6 A0A1A7YQB2 A0A1A8AV83 A0A1A8FS18 A0A1A8I9P4 Q6PGV3 A0A091UNW6 A0A1A8CZL3 H2SF11 Q6DE31 A0A3B4T610 A0A3P9IFX2 A0A3B4XWZ3 A0A3B3S904 A0A1S3AQG8 Q32NS1 A0A131Z1E1 A0A224Z1S4 A0A3P9LE43 U3I667 A0A3B3HTB1 A0A091L8S4 A0A0F8ASG4 A0A0F8CDY2 A0A2I0MUY5 T1J6X2 V8NTP9 A0A3P8Y1I9 A0A1V4JKN8 A0A0N8K2Y7 A0A091GRT6 A0A093SD50 A0A1S3PUM2 A0A1W4ZRP3 A0A091E759 A0A3B5QDC7 A0A087R6V8 A0A093C1C4 A0A1U7RML9 A0A093GW99 A0A3B4H5R0 A0A3P9BAP0 A0A093IHX6 A0A2I0TZE4 A0A093THY2 A0A087VMR2 A0A091JIJ6 A0A0A0AW15 A0A094K9L4 A0A091G7B1 A0A093PEU5 A0A094KPM5 A0A091PNY6 A0A091SDY0 A0A093I049 A0A218VAS9 A0A099YUS7 M7BA75 A0A3P8Q3L4 A0A091MCG8 U3JL60
PDB
6F3T
E-value=4.03928e-56,
Score=554
Ontologies
GO
PANTHER
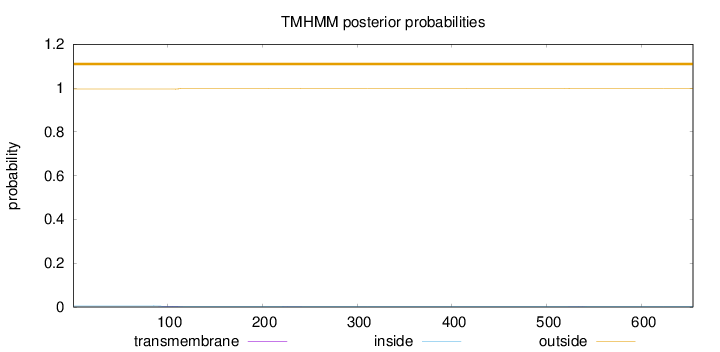
Topology
Length:
654
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06847
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00491
outside
1 - 654
Population Genetic Test Statistics
Pi
257.913755
Theta
166.056482
Tajima's D
1.702716
CLR
0.059705
CSRT
0.830708464576771
Interpretation
Uncertain
