Gene
KWMTBOMO03388
Annotation
reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.282 Nuclear Reliability : 1.076
Sequence
CDS
ATGAAGCTCGGTCGCGCTCCCGATTCCGTTCCCGTCACGAGGACTGTGGTGGATTGGCACACGCTGGGCATCAGCCTGGCTGAATCTGATCCACCATCGCTCCCGTTTAACCCGGACTCTATCCCGTCTCCTCAGGATACCGCTGAAGCCATAGACATCTTAACGTCACACATCACCTCGACATTAGATAGGTCATCGAAACAAGTTGTAGCGGAGGACTTCCTTCACCGCTTCAAATTGTCCGACGATATTAGGGAACTCCTTAGAGCTAAGAACGCCTCGATACGCGCCTACGACAGGTATCCTACCGCGGAAAATCGTATTCGAATGCGTGCCCTACAACGCGACGTAAAGTCTCGCATCGCCGAAGTCCGAGATGCCAGATGGTCTGATTTCTTAGAAGGACTCGCGCCCTCCCAAAGGTCTTACTACCGCTTAGCTCGTACTCTCAAATCGGATACGGTAGTAACTATGCCCCCCCTCGTAGGCCCCTCAGGCCGACTCGCGGCGTTCGATGATGACGAAAAAGCAGAGCTGCTGGCCGATACATTGCAAACCCAGTGCACGCCCAGCACTCAATCCGTGGACCCTGTTCATGTAGAATTAGTAGACAGTGAGGTAGAACGCAGAGCCTCCTTGCCACCATCGGATGCGTTACCACCCGTCACCCCGATAGAAGTTAAAGACTTGATCAAAGACCTACGTCCTCGCAAGGCTTCCGGTTCCGACGGTATATCTAACCGCGTTATTAAACTTCTACCCGTCCAACTCATCGTGATGTTGGCATCTATTTTCAATGCCGCTATGGCGAACTGTATCTTTCCCGCGGTGTGGAAAGAAGCGGACGTTATCGGCATACATAAACCCGGTAAACCAAAAAATCATTCGACGAGCTACCGCCCTATTAGCCTCCTCATGTCTCTAGGCAAACTGTATGAGCGTCTGCTCTACAAACGCCTCAGAGACTTCGTCTCATCCAAGGGCATTCTCATCGATGAACAATTCGGATTCCGTACAAATCACTCATGCGTTCAACAGGTGCACCGCCTCACGGAGCACATTCTTGTGGGGCTTAATCGACCAAAACCGTTATACACGGGAGCTCTCTTCTTCGACGTCGCAAAAGCGTTCGACAAAGTCTGGCACAATGGTTTGATTTTCAAACTATTCAACATGGGCGTGCCGGATAGTCTCGTGCTCATCATACGGGACTTCTTATCGAACCGCTCTTTTCGATATCGAGTCGAGGGAACCCGCTCCTCCCCACGACCTCTCACAGCTGGAGTCCCGCAAGGCTCTGTCCTTTCACCCCTCCTATTTAGCTTATTCGTCAACGATATTCCCCGGTCGCCGCCGACCCATTTAGCTTTATTCGCCGACGACACGACTGTTTACTATTCTAGTAGAAATAAGTCCCTAATCGCGAAGAAGCTTCAGAGCGCAGCCCTAGCCCTAGGACAGTGGTTCCGAAAATGGCGCATAGACATCAACCCAGCGAAATAG
Protein
MKLGRAPDSVPVTRTVVDWHTLGISLAESDPPSLPFNPDSIPSPQDTAEAIDILTSHITSTLDRSSKQVVAEDFLHRFKLSDDIRELLRAKNASIRAYDRYPTAENRIRMRALQRDVKSRIAEVRDARWSDFLEGLAPSQRSYYRLARTLKSDTVVTMPPLVGPSGRLAAFDDDEKAELLADTLQTQCTPSTQSVDPVHVELVDSEVERRASLPPSDALPPVTPIEVKDLIKDLRPRKASGSDGISNRVIKLLPVQLIVMLASIFNAAMANCIFPAVWKEADVIGIHKPGKPKNHSTSYRPISLLMSLGKLYERLLYKRLRDFVSSKGILIDEQFGFRTNHSCVQQVHRLTEHILVGLNRPKPLYTGALFFDVAKAFDKVWHNGLIFKLFNMGVPDSLVLIIRDFLSNRSFRYRVEGTRSSPRPLTAGVPQGSVLSPLLFSLFVNDIPRSPPTHLALFADDTTVYYSSRNKSLIAKKLQSAALALGQWFRKWRIDINPAK
Summary
Uniprot
Q93137
Q9XXW0
Q9BPQ0
A0A0N1PFV1
O96546
A0A1Y1NA23
+ More
A0A139W8G9 A0A139WA11 D7EM17 V5GNJ2 A0A2S2Q352 A0A2S2QEE0 A0A2S2PR14 K7IN67 V5GNF3 A0A2H8U0H5 A0A0C9RJA4 J9KKL3 A0A1B6J5Z9 A0A3N5Z7I3 J9JS95 X1WXE6 J9KLB1 A0A224XF24 A0A023EYS8 X1WLB2 A0A2S2PWA8 J9LC21 A0A2S2RAN7 A0A2S2PPK8 Q07995 A0A2S2R9D4 A0A2J7QUJ9 A0A2J7Q2R1 X1WN18 J9KCZ4 A0A2S2PBB4 J9KC83 J9KEU4 X1WZH0 A0A2J7R7D0 A0A2J7RPU2 A0A2S2NGV8 J9KPF1 A0A2J7QFL0 A0A2J7RKP3 A0A2J7R2E1 A0A2S2NJB5 A0A2J7PU96 A0A2J7RS77 X1WNI2 A0A2J7RE85 A0A2J7Q0Q6 A0A2S2PA10 A0A232ENU1 A0A087U3K8 V9H092 A0A2J7R6U1 A0A224XB44 A0A2J7QR37 A0A023F700 A0A2J7QY69 A0A2J7PBY3 A0A2S2PEZ9
A0A139W8G9 A0A139WA11 D7EM17 V5GNJ2 A0A2S2Q352 A0A2S2QEE0 A0A2S2PR14 K7IN67 V5GNF3 A0A2H8U0H5 A0A0C9RJA4 J9KKL3 A0A1B6J5Z9 A0A3N5Z7I3 J9JS95 X1WXE6 J9KLB1 A0A224XF24 A0A023EYS8 X1WLB2 A0A2S2PWA8 J9LC21 A0A2S2RAN7 A0A2S2PPK8 Q07995 A0A2S2R9D4 A0A2J7QUJ9 A0A2J7Q2R1 X1WN18 J9KCZ4 A0A2S2PBB4 J9KC83 J9KEU4 X1WZH0 A0A2J7R7D0 A0A2J7RPU2 A0A2S2NGV8 J9KPF1 A0A2J7QFL0 A0A2J7RKP3 A0A2J7R2E1 A0A2S2NJB5 A0A2J7PU96 A0A2J7RS77 X1WNI2 A0A2J7RE85 A0A2J7Q0Q6 A0A2S2PA10 A0A232ENU1 A0A087U3K8 V9H092 A0A2J7R6U1 A0A224XB44 A0A2J7QR37 A0A023F700 A0A2J7QY69 A0A2J7PBY3 A0A2S2PEZ9
Pubmed
EMBL
U07847
AAA17752.1
AB018558
BAA76304.1
AB055391
BAB21761.1
+ More
KQ459265 KPJ02064.1 AF081103 AAC72921.1 GEZM01008457 JAV94772.1 KQ973342 KXZ75569.1 KQ971729 KYB24751.1 KQ973412 EFA12510.2 GALX01005299 JAB63167.1 GGMS01002955 MBY72158.1 GGMS01006359 MBY75562.1 GGMR01019198 MBY31817.1 GALX01005359 JAB63107.1 GFXV01007093 MBW18898.1 GBYB01008290 JAG78057.1 ABLF02029306 ABLF02029314 ABLF02060138 GECU01013097 JAS94609.1 RPOV01000134 RPJ78669.1 ABLF02028535 ABLF02028545 ABLF02016634 ABLF02040174 ABLF02040183 ABLF02055266 GFTR01008038 JAW08388.1 GBBI01004395 JAC14317.1 ABLF02042518 ABLF02057746 GGMS01000625 MBY69828.1 ABLF02036316 ABLF02036319 ABLF02048663 ABLF02066977 GGMS01017914 MBY87117.1 GGMR01018716 MBY31335.1 S59870 AAB26437.2 GGMS01017365 MBY86568.1 NEVH01010546 PNF32261.1 NEVH01019080 PNF22876.1 ABLF02021177 ABLF02027367 ABLF02066338 ABLF02008463 GGMR01014053 MBY26672.1 ABLF02033757 ABLF02040650 ABLF02040651 ABLF02056776 ABLF02034467 NEVH01006736 PNF36716.1 NEVH01001355 PNF42855.1 GGMR01003407 MBY16026.1 ABLF02041767 NEVH01014858 PNF27387.1 NEVH01002716 PNF41415.1 NEVH01007838 PNF34986.1 GGMR01004661 MBY17280.1 NEVH01021205 PNF19912.1 NEVH01000280 PNF43672.1 ABLF02008153 ABLF02008462 ABLF02008464 NEVH01005277 PNF39135.1 NEVH01019964 PNF22161.1 GGMR01013409 MBY26028.1 NNAY01003095 OXU19992.1 KK118005 KFM71947.1 L79944 AAB04627.1 NEVH01006754 PNF36554.1 GFTR01008262 JAW08164.1 NEVH01011985 PNF31054.1 GBBI01001699 JAC17013.1 NEVH01009134 PNF33528.1 NEVH01027074 PNF13841.1 GGMR01015412 MBY28031.1
KQ459265 KPJ02064.1 AF081103 AAC72921.1 GEZM01008457 JAV94772.1 KQ973342 KXZ75569.1 KQ971729 KYB24751.1 KQ973412 EFA12510.2 GALX01005299 JAB63167.1 GGMS01002955 MBY72158.1 GGMS01006359 MBY75562.1 GGMR01019198 MBY31817.1 GALX01005359 JAB63107.1 GFXV01007093 MBW18898.1 GBYB01008290 JAG78057.1 ABLF02029306 ABLF02029314 ABLF02060138 GECU01013097 JAS94609.1 RPOV01000134 RPJ78669.1 ABLF02028535 ABLF02028545 ABLF02016634 ABLF02040174 ABLF02040183 ABLF02055266 GFTR01008038 JAW08388.1 GBBI01004395 JAC14317.1 ABLF02042518 ABLF02057746 GGMS01000625 MBY69828.1 ABLF02036316 ABLF02036319 ABLF02048663 ABLF02066977 GGMS01017914 MBY87117.1 GGMR01018716 MBY31335.1 S59870 AAB26437.2 GGMS01017365 MBY86568.1 NEVH01010546 PNF32261.1 NEVH01019080 PNF22876.1 ABLF02021177 ABLF02027367 ABLF02066338 ABLF02008463 GGMR01014053 MBY26672.1 ABLF02033757 ABLF02040650 ABLF02040651 ABLF02056776 ABLF02034467 NEVH01006736 PNF36716.1 NEVH01001355 PNF42855.1 GGMR01003407 MBY16026.1 ABLF02041767 NEVH01014858 PNF27387.1 NEVH01002716 PNF41415.1 NEVH01007838 PNF34986.1 GGMR01004661 MBY17280.1 NEVH01021205 PNF19912.1 NEVH01000280 PNF43672.1 ABLF02008153 ABLF02008462 ABLF02008464 NEVH01005277 PNF39135.1 NEVH01019964 PNF22161.1 GGMR01013409 MBY26028.1 NNAY01003095 OXU19992.1 KK118005 KFM71947.1 L79944 AAB04627.1 NEVH01006754 PNF36554.1 GFTR01008262 JAW08164.1 NEVH01011985 PNF31054.1 GBBI01001699 JAC17013.1 NEVH01009134 PNF33528.1 NEVH01027074 PNF13841.1 GGMR01015412 MBY28031.1
Proteomes
Pfam
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
Q93137
Q9XXW0
Q9BPQ0
A0A0N1PFV1
O96546
A0A1Y1NA23
+ More
A0A139W8G9 A0A139WA11 D7EM17 V5GNJ2 A0A2S2Q352 A0A2S2QEE0 A0A2S2PR14 K7IN67 V5GNF3 A0A2H8U0H5 A0A0C9RJA4 J9KKL3 A0A1B6J5Z9 A0A3N5Z7I3 J9JS95 X1WXE6 J9KLB1 A0A224XF24 A0A023EYS8 X1WLB2 A0A2S2PWA8 J9LC21 A0A2S2RAN7 A0A2S2PPK8 Q07995 A0A2S2R9D4 A0A2J7QUJ9 A0A2J7Q2R1 X1WN18 J9KCZ4 A0A2S2PBB4 J9KC83 J9KEU4 X1WZH0 A0A2J7R7D0 A0A2J7RPU2 A0A2S2NGV8 J9KPF1 A0A2J7QFL0 A0A2J7RKP3 A0A2J7R2E1 A0A2S2NJB5 A0A2J7PU96 A0A2J7RS77 X1WNI2 A0A2J7RE85 A0A2J7Q0Q6 A0A2S2PA10 A0A232ENU1 A0A087U3K8 V9H092 A0A2J7R6U1 A0A224XB44 A0A2J7QR37 A0A023F700 A0A2J7QY69 A0A2J7PBY3 A0A2S2PEZ9
A0A139W8G9 A0A139WA11 D7EM17 V5GNJ2 A0A2S2Q352 A0A2S2QEE0 A0A2S2PR14 K7IN67 V5GNF3 A0A2H8U0H5 A0A0C9RJA4 J9KKL3 A0A1B6J5Z9 A0A3N5Z7I3 J9JS95 X1WXE6 J9KLB1 A0A224XF24 A0A023EYS8 X1WLB2 A0A2S2PWA8 J9LC21 A0A2S2RAN7 A0A2S2PPK8 Q07995 A0A2S2R9D4 A0A2J7QUJ9 A0A2J7Q2R1 X1WN18 J9KCZ4 A0A2S2PBB4 J9KC83 J9KEU4 X1WZH0 A0A2J7R7D0 A0A2J7RPU2 A0A2S2NGV8 J9KPF1 A0A2J7QFL0 A0A2J7RKP3 A0A2J7R2E1 A0A2S2NJB5 A0A2J7PU96 A0A2J7RS77 X1WNI2 A0A2J7RE85 A0A2J7Q0Q6 A0A2S2PA10 A0A232ENU1 A0A087U3K8 V9H092 A0A2J7R6U1 A0A224XB44 A0A2J7QR37 A0A023F700 A0A2J7QY69 A0A2J7PBY3 A0A2S2PEZ9
Ontologies
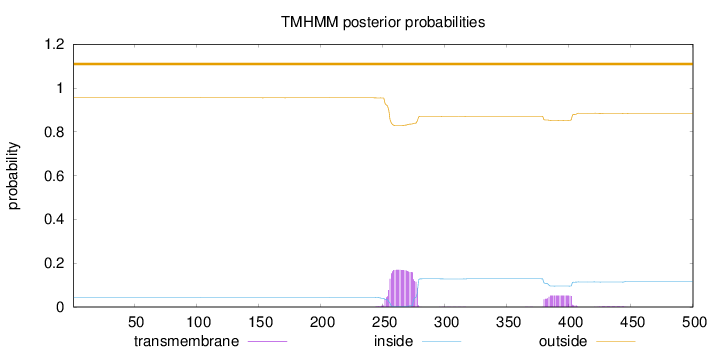
Topology
Length:
500
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.04051
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04468
outside
1 - 500
Population Genetic Test Statistics
Pi
0.173544
Theta
0.160893
Tajima's D
0
CLR
135.165055
CSRT
0.227038648067597
Interpretation
Uncertain
