Gene
KWMTBOMO03381
Pre Gene Modal
BGIBMGA006384
Annotation
abdominal_B_isoform_RBL_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.551
Sequence
CDS
ATGATGAACGGGGTCGGCGGCGCGCTCTACGAGGAGGCGCACGCGTCGCCGCCCGGTGGCTCACCCCCAGCAGCTCCTGCGGCCGCACCACCTTCGGCCTCCAGCGCCTCGCCCGCTTCAGTCGGCTCCAACCCACCACAACCGCCGTTGCACATTCCGGCTAAGCGCTACGAGCCCGAGCCCGGCGTCATCCGGCATGCTCAGCAGCCCTGGGGCTATCCTCCAGATGCTCCCGGCTCATTCGAACATCAGTACCCCAGCGCCCCTACATATTATAATTTACCTGTCGAAAGAGAGCGCAAGTCGACTCTCCCCTTTTGGCCAACCAGCAGTGGTGAATACAAACCATACGCTGATGGAGGCTGTCATCAGGGTTTCTCGCAGCCCTGCTGGAACTATCCGTATGGCACACCTAGAGGCGATCAACCTCTGCCCTACGTCAGCGGAGAAGAACGACGAGCTGCAGTCTCCGAGGCGTCCGGGTTCTCTCATGATGCATATGGTCTACGAAATTACGCACCAGAGTCAGTTTCTGGCGCACCGTATCCGCCACCAGGTTCTCTACCCGGTTCTCTGTCAATGTCGGTCGGAGTCGGGGTGGGGTGTGGATCCTCGAACCCTCTGGATTGGACTGGTCAAGTGACGGTGCGCAAAAAACGCAAACCATACTCGAAATTTCAAACTCTCGAACTGGAAAAGGAATTCCTTTTCAACGCAATAGACGAATGA
Protein
MMNGVGGALYEEAHASPPGGSPPAAPAAAPPSASSASPASVGSNPPQPPLHIPAKRYEPEPGVIRHAQQPWGYPPDAPGSFEHQYPSAPTYYNLPVERERKSTLPFWPTSSGEYKPYADGGCHQGFSQPCWNYPYGTPRGDQPLPYVSGEERRAAVSEASGFSHDAYGLRNYAPESVSGAPYPPPGSLPGSLSMSVGVGVGCGSSNPLDWTGQVTVRKKRKPYSKFQTLELEKEFLFNAIDE
Summary
Uniprot
B9ZZS7
B9ZZS8
A0A2H1VL22
A0A2A4JSK0
A0A0L7LHS9
A0A194Q313
+ More
A0A0N1IEG1 A0A1Q1M8Y2 H9JA39 A0A224XSF5 E0VI18 A0A139WMT7 Q9NHB6 A0A2R7W7V4 A0A0A9YMH5 N6TLW2 U4UJ34 A0A1B6CEF2 A0A1Y1LAQ2 A0A059PB59 A0A059PBA2 A0A0M3MZ45 A0A067RHN3 A0A1B6MDF5 Q1A1Y3 A0A1Y1LC97 A0A0M9A761 A0A088AID8 Q1KY82 A0A154P7C9 E2B5X1 T1IST5 Q1A1Y4 A0A0C9PW24 A0A3L8DCK4 A0A2A3EJP3 Q9XZR3 A0A026W8B4 A0A087T6M5 K7J1B1 E2AHB3 A0A0K8VZ57 A0A0K8UPT8 A0A0K8VK32 U3UBU1 A0A3B0JQF7 B4NJG3 A0A1D2NBP1 I5AP43 B4G4I3 A0A1W4VV26 A0A034W194 A0A0A1XGN6 W8BHX8
A0A0N1IEG1 A0A1Q1M8Y2 H9JA39 A0A224XSF5 E0VI18 A0A139WMT7 Q9NHB6 A0A2R7W7V4 A0A0A9YMH5 N6TLW2 U4UJ34 A0A1B6CEF2 A0A1Y1LAQ2 A0A059PB59 A0A059PBA2 A0A0M3MZ45 A0A067RHN3 A0A1B6MDF5 Q1A1Y3 A0A1Y1LC97 A0A0M9A761 A0A088AID8 Q1KY82 A0A154P7C9 E2B5X1 T1IST5 Q1A1Y4 A0A0C9PW24 A0A3L8DCK4 A0A2A3EJP3 Q9XZR3 A0A026W8B4 A0A087T6M5 K7J1B1 E2AHB3 A0A0K8VZ57 A0A0K8UPT8 A0A0K8VK32 U3UBU1 A0A3B0JQF7 B4NJG3 A0A1D2NBP1 I5AP43 B4G4I3 A0A1W4VV26 A0A034W194 A0A0A1XGN6 W8BHX8
Pubmed
EMBL
AB461857
BAH30151.1
AB461858
BAH30152.1
ODYU01003142
SOQ41523.1
+ More
NWSH01000693 PCG74746.1 JTDY01001019 KOB75113.1 KQ459551 KPI99947.1 KQ460544 KPJ14090.1 KU663673 AQM32555.1 BABH01009207 GFTR01005517 JAW10909.1 DS235181 EEB13024.1 KQ971312 KYB29151.1 AF227923 AAF36721.1 EEZ99247.1 KK854294 PTY14285.1 GBHO01012889 GDHC01005462 JAG30715.1 JAQ13167.1 APGK01030460 KB740773 ENN78973.1 KB632345 ERL93097.1 GEDC01028792 GEDC01025598 JAS08506.1 JAS11700.1 GEZM01062220 GEZM01062219 GEZM01062215 JAV69948.1 JQ952575 AGC12524.1 JQ952574 AGC12523.1 KM496286 AKM12294.1 KK852676 KDR18691.1 GEBQ01006021 JAT33956.1 DQ383821 ABD38146.1 GEZM01062217 GEZM01062216 GEZM01062213 JAV69950.1 KQ435720 KOX78625.1 DQ368691 ABD16214.1 KQ434820 KZC07030.1 GL445887 EFN88940.1 JH431446 DQ383820 ABD38145.1 GBYB01005693 JAG75460.1 QOIP01000010 RLU18061.1 KZ288222 PBC32033.1 AJ131397 CAB40807.1 KK107348 EZA52203.1 KK113668 KFM60764.1 GL439483 EFN67230.1 GDHF01008459 JAI43855.1 GDHF01023718 JAI28596.1 GDHF01013374 JAI38940.1 HE979845 CCK73379.1 OUUW01000007 SPP83183.1 CH964272 EDW83887.2 LJIJ01000100 ODN02667.1 CM000070 EIM52728.2 CH479179 EDW25154.1 GAKP01009626 JAC49326.1 GBXI01004212 JAD10080.1 GAMC01013704 JAB92851.1
NWSH01000693 PCG74746.1 JTDY01001019 KOB75113.1 KQ459551 KPI99947.1 KQ460544 KPJ14090.1 KU663673 AQM32555.1 BABH01009207 GFTR01005517 JAW10909.1 DS235181 EEB13024.1 KQ971312 KYB29151.1 AF227923 AAF36721.1 EEZ99247.1 KK854294 PTY14285.1 GBHO01012889 GDHC01005462 JAG30715.1 JAQ13167.1 APGK01030460 KB740773 ENN78973.1 KB632345 ERL93097.1 GEDC01028792 GEDC01025598 JAS08506.1 JAS11700.1 GEZM01062220 GEZM01062219 GEZM01062215 JAV69948.1 JQ952575 AGC12524.1 JQ952574 AGC12523.1 KM496286 AKM12294.1 KK852676 KDR18691.1 GEBQ01006021 JAT33956.1 DQ383821 ABD38146.1 GEZM01062217 GEZM01062216 GEZM01062213 JAV69950.1 KQ435720 KOX78625.1 DQ368691 ABD16214.1 KQ434820 KZC07030.1 GL445887 EFN88940.1 JH431446 DQ383820 ABD38145.1 GBYB01005693 JAG75460.1 QOIP01000010 RLU18061.1 KZ288222 PBC32033.1 AJ131397 CAB40807.1 KK107348 EZA52203.1 KK113668 KFM60764.1 GL439483 EFN67230.1 GDHF01008459 JAI43855.1 GDHF01023718 JAI28596.1 GDHF01013374 JAI38940.1 HE979845 CCK73379.1 OUUW01000007 SPP83183.1 CH964272 EDW83887.2 LJIJ01000100 ODN02667.1 CM000070 EIM52728.2 CH479179 EDW25154.1 GAKP01009626 JAC49326.1 GBXI01004212 JAD10080.1 GAMC01013704 JAB92851.1
Proteomes
UP000218220
UP000037510
UP000053268
UP000053240
UP000005204
UP000009046
+ More
UP000007266 UP000019118 UP000030742 UP000027135 UP000053105 UP000005203 UP000076502 UP000008237 UP000279307 UP000242457 UP000053097 UP000054359 UP000002358 UP000000311 UP000268350 UP000007798 UP000094527 UP000001819 UP000008744 UP000192221
UP000007266 UP000019118 UP000030742 UP000027135 UP000053105 UP000005203 UP000076502 UP000008237 UP000279307 UP000242457 UP000053097 UP000054359 UP000002358 UP000000311 UP000268350 UP000007798 UP000094527 UP000001819 UP000008744 UP000192221
Pfam
PF00046 Homeodomain
Interpro
SUPFAM
SSF46689
SSF46689
ProteinModelPortal
B9ZZS7
B9ZZS8
A0A2H1VL22
A0A2A4JSK0
A0A0L7LHS9
A0A194Q313
+ More
A0A0N1IEG1 A0A1Q1M8Y2 H9JA39 A0A224XSF5 E0VI18 A0A139WMT7 Q9NHB6 A0A2R7W7V4 A0A0A9YMH5 N6TLW2 U4UJ34 A0A1B6CEF2 A0A1Y1LAQ2 A0A059PB59 A0A059PBA2 A0A0M3MZ45 A0A067RHN3 A0A1B6MDF5 Q1A1Y3 A0A1Y1LC97 A0A0M9A761 A0A088AID8 Q1KY82 A0A154P7C9 E2B5X1 T1IST5 Q1A1Y4 A0A0C9PW24 A0A3L8DCK4 A0A2A3EJP3 Q9XZR3 A0A026W8B4 A0A087T6M5 K7J1B1 E2AHB3 A0A0K8VZ57 A0A0K8UPT8 A0A0K8VK32 U3UBU1 A0A3B0JQF7 B4NJG3 A0A1D2NBP1 I5AP43 B4G4I3 A0A1W4VV26 A0A034W194 A0A0A1XGN6 W8BHX8
A0A0N1IEG1 A0A1Q1M8Y2 H9JA39 A0A224XSF5 E0VI18 A0A139WMT7 Q9NHB6 A0A2R7W7V4 A0A0A9YMH5 N6TLW2 U4UJ34 A0A1B6CEF2 A0A1Y1LAQ2 A0A059PB59 A0A059PBA2 A0A0M3MZ45 A0A067RHN3 A0A1B6MDF5 Q1A1Y3 A0A1Y1LC97 A0A0M9A761 A0A088AID8 Q1KY82 A0A154P7C9 E2B5X1 T1IST5 Q1A1Y4 A0A0C9PW24 A0A3L8DCK4 A0A2A3EJP3 Q9XZR3 A0A026W8B4 A0A087T6M5 K7J1B1 E2AHB3 A0A0K8VZ57 A0A0K8UPT8 A0A0K8VK32 U3UBU1 A0A3B0JQF7 B4NJG3 A0A1D2NBP1 I5AP43 B4G4I3 A0A1W4VV26 A0A034W194 A0A0A1XGN6 W8BHX8
PDB
5ZJT
E-value=1.41119e-09,
Score=148
Ontologies
KEGG
GO
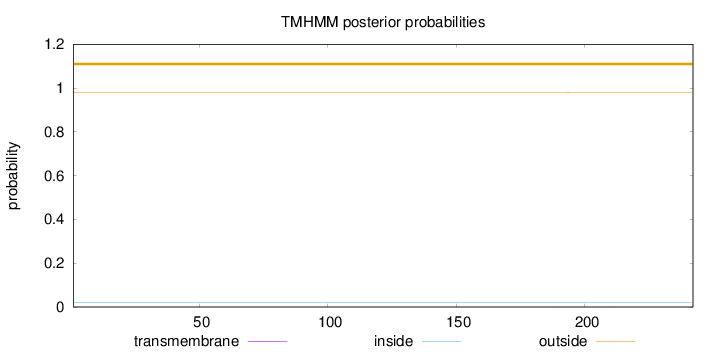
Topology
Subcellular location
Nucleus
Length:
242
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00923
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02034
outside
1 - 242
Population Genetic Test Statistics
Pi
213.04618
Theta
208.149665
Tajima's D
0.101103
CLR
17.979366
CSRT
0.398430078496075
Interpretation
Uncertain
