Pre Gene Modal
BGIBMGA006401
Annotation
PREDICTED:_glutaredoxin_3_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 3.354
Sequence
CDS
ATGTCGGTCAGTAAATTGGACTCTGTTGAAGATTTTAATAATTTTATAAGATCACCTGCACTTGGAGTAGTACACTTTTCAGCAGAATGGGCCGAACAATGTAAACAAGTTACAGATGTACTTGAAGAACTCTTAAAACTTCCTGAAATTCAGTCGAGCAAGACACAATGTGCTGTTTGTGATGCCGAGGCACTTTCTGAAGTTTCCTTGCAATATAAGGTAGATTCAGTGCCTACAGTTATATTATTTAAGAATGGAACTCAAGTAGACAGGATTGATGGAGCAGATGCTGCTCAAATTAGCACAAAAATTAAAGCACAGAGTCTTAATAAGAGTCCTGCAGAAATAACCCCTCAGAAACTTGAAGATCGACTAAAAGCTTTAATAAATAAGCATAATATAATGGTATTCATGAAAGGGAACAAGGAAACTCCAAGATGCGGGTTTAGTCGTACTCTCATACAAATTCTGAATGGAACAGGCGTTCAATTTGGTACTTTTGACATATTGACGGATGAAGAGGTTCGTCAGGGTCTGAAAGAGTATTCTGATTGGCCAACGTATCCTCAGCTTTATGTAAAAGGAGAACTGATTGGAGGTTTGGATATAATAAAAGAATTACAAGCCAATGGGGAGCTAGAAGCAACACTAAACCCATAA
Protein
MSVSKLDSVEDFNNFIRSPALGVVHFSAEWAEQCKQVTDVLEELLKLPEIQSSKTQCAVCDAEALSEVSLQYKVDSVPTVILFKNGTQVDRIDGADAAQISTKIKAQSLNKSPAEITPQKLEDRLKALINKHNIMVFMKGNKETPRCGFSRTLIQILNGTGVQFGTFDILTDEEVRQGLKEYSDWPTYPQLYVKGELIGGLDIIKELQANGELEATLNP
Summary
Uniprot
H9JA56
A0A1N7THB0
A0A2A4JEH4
A0A2H1WA34
S4PS39
A0A212F1K1
+ More
A0A194Q971 A0A194RAI7 A0A0L7LBG6 A0A0T6AV65 D6WB96 A0A1Q3G293 Q17G46 A0A023EJW2 N6SY01 J3JX56 B4MWH2 A0A182TL42 A0A182XK69 A0A182KWY9 Q7QDJ9 A0A182HJR9 A0A182V8K1 A0A182K0J1 A0A2M3Z6P3 A0A1W4WZZ6 W5JPB3 A0A182IK21 T1E7U8 A0A182NKV0 A0A2M4BZ11 A0A084VZD2 A0A1I8M1A7 A0A1Y1MVF5 Q9VJZ6 A0A2M3Z6S5 A0A182R811 A0A2M4AQ69 A0A182QLC2 A0A2M4APD1 W8CDM7 A0A1L8ED64 A0A1L8DU36 V5H0Y1 A0A1I8Q318 A0A2M4APH6 A0A3B0KPJ4 B4JP71 A0A182PS52 A0A1B0D3Y8 B4GKA8 A0A034V9R6 Q29P85 A0A0K8WEH2 A0A1W4V8N9 U4U4Z2 B4KK55 B4Q4S2 A0A0M5J1Z2 B3NAY1 B4M976 A0A0L0C211 A0A182SHE2 A0A182LRT3 B4P353 A0A1A9XRQ8 A0A182W0K3 A0A1A9W8I7 B3MV87 B4IEK1 A0A336LI50 A0A0L7RJM1 A0A0M8ZZD8 E2APR1 A0A2A3EF26 A0A088ADE6 A0A0B7AJV3 A0A026X5R4 A0A3L8DJ19 A0A0K8TTB2 A0A0B7AJI6 A0A154NW11 A0A1B6FH87 A0A0J7L395 A0A0P4WFF9 A0A158NVI5 E0VTK1 A0A2H4D133 A0A195BN03 A0A1B6ISW9 V4A538 F4WLF4 K7IVE7 A0A232EV28 A0A2J7RFU8 A0A3B3DW59 A0A067QVZ4 A0A1W4Y1D7 A0A2A2L411 A0A2H8TR00
A0A194Q971 A0A194RAI7 A0A0L7LBG6 A0A0T6AV65 D6WB96 A0A1Q3G293 Q17G46 A0A023EJW2 N6SY01 J3JX56 B4MWH2 A0A182TL42 A0A182XK69 A0A182KWY9 Q7QDJ9 A0A182HJR9 A0A182V8K1 A0A182K0J1 A0A2M3Z6P3 A0A1W4WZZ6 W5JPB3 A0A182IK21 T1E7U8 A0A182NKV0 A0A2M4BZ11 A0A084VZD2 A0A1I8M1A7 A0A1Y1MVF5 Q9VJZ6 A0A2M3Z6S5 A0A182R811 A0A2M4AQ69 A0A182QLC2 A0A2M4APD1 W8CDM7 A0A1L8ED64 A0A1L8DU36 V5H0Y1 A0A1I8Q318 A0A2M4APH6 A0A3B0KPJ4 B4JP71 A0A182PS52 A0A1B0D3Y8 B4GKA8 A0A034V9R6 Q29P85 A0A0K8WEH2 A0A1W4V8N9 U4U4Z2 B4KK55 B4Q4S2 A0A0M5J1Z2 B3NAY1 B4M976 A0A0L0C211 A0A182SHE2 A0A182LRT3 B4P353 A0A1A9XRQ8 A0A182W0K3 A0A1A9W8I7 B3MV87 B4IEK1 A0A336LI50 A0A0L7RJM1 A0A0M8ZZD8 E2APR1 A0A2A3EF26 A0A088ADE6 A0A0B7AJV3 A0A026X5R4 A0A3L8DJ19 A0A0K8TTB2 A0A0B7AJI6 A0A154NW11 A0A1B6FH87 A0A0J7L395 A0A0P4WFF9 A0A158NVI5 E0VTK1 A0A2H4D133 A0A195BN03 A0A1B6ISW9 V4A538 F4WLF4 K7IVE7 A0A232EV28 A0A2J7RFU8 A0A3B3DW59 A0A067QVZ4 A0A1W4Y1D7 A0A2A2L411 A0A2H8TR00
Pubmed
19121390
23622113
22118469
26354079
26227816
18362917
+ More
19820115 17510324 24945155 26483478 23537049 22516182 17994087 20966253 12364791 14747013 17210077 20920257 23761445 24438588 25315136 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 25348373 15632085 22936249 26108605 17550304 20798317 24508170 30249741 26369729 21347285 20566863 23254933 21719571 20075255 28648823 29451363 24845553
19820115 17510324 24945155 26483478 23537049 22516182 17994087 20966253 12364791 14747013 17210077 20920257 23761445 24438588 25315136 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 25348373 15632085 22936249 26108605 17550304 20798317 24508170 30249741 26369729 21347285 20566863 23254933 21719571 20075255 28648823 29451363 24845553
EMBL
BABH01009116
BABH01009117
KU356684
AMW64459.1
NWSH01001882
PCG69802.1
+ More
ODYU01006994 SOQ49344.1 GAIX01000260 JAA92300.1 AGBW02010865 OWR47606.1 KQ459551 KPI99970.1 KQ460473 KPJ14514.1 JTDY01001806 KOB72832.1 LJIG01022740 KRT78961.1 KQ971312 EEZ97918.1 GFDL01001110 JAV33935.1 CH477266 EAT45539.1 JXUM01060983 GAPW01004221 KQ562129 JAC09377.1 KXJ76623.1 APGK01052662 KB741213 ENN72644.1 BT127824 AEE62786.1 CH963857 EDW76113.1 AAAB01008851 EAA07378.2 APCN01002207 GGFM01003445 MBW24196.1 ADMH02000756 ETN65158.1 GAMD01002737 JAA98853.1 GGFJ01009159 MBW58300.1 ATLV01018745 KE525248 KFB43326.1 GEZM01019946 GEZM01019945 GEZM01019944 GEZM01019943 JAV89529.1 AE014134 AY061456 AAF53288.1 AAL29004.1 GGFM01003478 MBW24229.1 GGFK01009437 MBW42758.1 AXCN02000306 GGFK01009323 MBW42644.1 GAMC01001676 JAC04880.1 GFDG01002207 JAV16592.1 GFDF01004133 JAV09951.1 GALX01001978 JAB66488.1 GGFK01009363 MBW42684.1 OUUW01000010 SPP85768.1 CH916372 EDV98701.1 AJVK01023814 CH479184 EDW37074.1 GAKP01020112 GAKP01020105 GAKP01020103 GAKP01020100 JAC38847.1 CH379058 EAL34408.1 GDHF01002812 JAI49502.1 KB631653 ERL85040.1 CH933807 EDW11573.1 CM000361 CM002910 EDX04910.1 KMY90081.1 CP012523 ALC39961.1 CH954177 EDV58695.1 CH940654 EDW57752.1 JRES01000996 KNC26306.1 AXCM01000236 CM000157 EDW88295.1 CH902624 EDV33152.1 CH480831 EDW46028.1 UFQS01004415 UFQT01004415 SSX16365.1 SSX35682.1 KQ414581 KOC71034.1 KQ435812 KOX72723.1 GL441581 EFN64537.1 KZ288271 PBC29809.1 HACG01033340 CEK80205.1 KK107019 EZA62809.1 QOIP01000007 RLU20407.1 GDAI01000443 JAI17160.1 HACG01033341 CEK80206.1 KQ434766 KZC03742.1 GECZ01020193 JAS49576.1 LBMM01000996 KMQ97093.1 GDRN01061136 JAI65246.1 ADTU01027203 DS235767 EEB16728.1 KU351385 AMA01982.1 KQ976433 KYM87360.1 GECU01017723 JAS89983.1 KB201194 ESO99023.1 GL888207 EGI65117.1 AAZX01004302 NNAY01002059 OXU22210.1 NEVH01004410 PNF39698.1 KK853305 KDR08711.1 LIAE01007226 PAV80855.1 GFXV01004819 MBW16624.1
ODYU01006994 SOQ49344.1 GAIX01000260 JAA92300.1 AGBW02010865 OWR47606.1 KQ459551 KPI99970.1 KQ460473 KPJ14514.1 JTDY01001806 KOB72832.1 LJIG01022740 KRT78961.1 KQ971312 EEZ97918.1 GFDL01001110 JAV33935.1 CH477266 EAT45539.1 JXUM01060983 GAPW01004221 KQ562129 JAC09377.1 KXJ76623.1 APGK01052662 KB741213 ENN72644.1 BT127824 AEE62786.1 CH963857 EDW76113.1 AAAB01008851 EAA07378.2 APCN01002207 GGFM01003445 MBW24196.1 ADMH02000756 ETN65158.1 GAMD01002737 JAA98853.1 GGFJ01009159 MBW58300.1 ATLV01018745 KE525248 KFB43326.1 GEZM01019946 GEZM01019945 GEZM01019944 GEZM01019943 JAV89529.1 AE014134 AY061456 AAF53288.1 AAL29004.1 GGFM01003478 MBW24229.1 GGFK01009437 MBW42758.1 AXCN02000306 GGFK01009323 MBW42644.1 GAMC01001676 JAC04880.1 GFDG01002207 JAV16592.1 GFDF01004133 JAV09951.1 GALX01001978 JAB66488.1 GGFK01009363 MBW42684.1 OUUW01000010 SPP85768.1 CH916372 EDV98701.1 AJVK01023814 CH479184 EDW37074.1 GAKP01020112 GAKP01020105 GAKP01020103 GAKP01020100 JAC38847.1 CH379058 EAL34408.1 GDHF01002812 JAI49502.1 KB631653 ERL85040.1 CH933807 EDW11573.1 CM000361 CM002910 EDX04910.1 KMY90081.1 CP012523 ALC39961.1 CH954177 EDV58695.1 CH940654 EDW57752.1 JRES01000996 KNC26306.1 AXCM01000236 CM000157 EDW88295.1 CH902624 EDV33152.1 CH480831 EDW46028.1 UFQS01004415 UFQT01004415 SSX16365.1 SSX35682.1 KQ414581 KOC71034.1 KQ435812 KOX72723.1 GL441581 EFN64537.1 KZ288271 PBC29809.1 HACG01033340 CEK80205.1 KK107019 EZA62809.1 QOIP01000007 RLU20407.1 GDAI01000443 JAI17160.1 HACG01033341 CEK80206.1 KQ434766 KZC03742.1 GECZ01020193 JAS49576.1 LBMM01000996 KMQ97093.1 GDRN01061136 JAI65246.1 ADTU01027203 DS235767 EEB16728.1 KU351385 AMA01982.1 KQ976433 KYM87360.1 GECU01017723 JAS89983.1 KB201194 ESO99023.1 GL888207 EGI65117.1 AAZX01004302 NNAY01002059 OXU22210.1 NEVH01004410 PNF39698.1 KK853305 KDR08711.1 LIAE01007226 PAV80855.1 GFXV01004819 MBW16624.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000037510
+ More
UP000007266 UP000008820 UP000069940 UP000249989 UP000019118 UP000007798 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075903 UP000075881 UP000192223 UP000000673 UP000075880 UP000075884 UP000030765 UP000095301 UP000000803 UP000075900 UP000075886 UP000095300 UP000268350 UP000001070 UP000075885 UP000092462 UP000008744 UP000001819 UP000192221 UP000030742 UP000009192 UP000000304 UP000092553 UP000008711 UP000008792 UP000037069 UP000075901 UP000075883 UP000002282 UP000092443 UP000075920 UP000091820 UP000007801 UP000001292 UP000053825 UP000053105 UP000000311 UP000242457 UP000005203 UP000053097 UP000279307 UP000076502 UP000036403 UP000005205 UP000009046 UP000078540 UP000030746 UP000007755 UP000002358 UP000215335 UP000235965 UP000261560 UP000027135 UP000192224 UP000218231
UP000007266 UP000008820 UP000069940 UP000249989 UP000019118 UP000007798 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075903 UP000075881 UP000192223 UP000000673 UP000075880 UP000075884 UP000030765 UP000095301 UP000000803 UP000075900 UP000075886 UP000095300 UP000268350 UP000001070 UP000075885 UP000092462 UP000008744 UP000001819 UP000192221 UP000030742 UP000009192 UP000000304 UP000092553 UP000008711 UP000008792 UP000037069 UP000075901 UP000075883 UP000002282 UP000092443 UP000075920 UP000091820 UP000007801 UP000001292 UP000053825 UP000053105 UP000000311 UP000242457 UP000005203 UP000053097 UP000279307 UP000076502 UP000036403 UP000005205 UP000009046 UP000078540 UP000030746 UP000007755 UP000002358 UP000215335 UP000235965 UP000261560 UP000027135 UP000192224 UP000218231
Interpro
Gene 3D
ProteinModelPortal
H9JA56
A0A1N7THB0
A0A2A4JEH4
A0A2H1WA34
S4PS39
A0A212F1K1
+ More
A0A194Q971 A0A194RAI7 A0A0L7LBG6 A0A0T6AV65 D6WB96 A0A1Q3G293 Q17G46 A0A023EJW2 N6SY01 J3JX56 B4MWH2 A0A182TL42 A0A182XK69 A0A182KWY9 Q7QDJ9 A0A182HJR9 A0A182V8K1 A0A182K0J1 A0A2M3Z6P3 A0A1W4WZZ6 W5JPB3 A0A182IK21 T1E7U8 A0A182NKV0 A0A2M4BZ11 A0A084VZD2 A0A1I8M1A7 A0A1Y1MVF5 Q9VJZ6 A0A2M3Z6S5 A0A182R811 A0A2M4AQ69 A0A182QLC2 A0A2M4APD1 W8CDM7 A0A1L8ED64 A0A1L8DU36 V5H0Y1 A0A1I8Q318 A0A2M4APH6 A0A3B0KPJ4 B4JP71 A0A182PS52 A0A1B0D3Y8 B4GKA8 A0A034V9R6 Q29P85 A0A0K8WEH2 A0A1W4V8N9 U4U4Z2 B4KK55 B4Q4S2 A0A0M5J1Z2 B3NAY1 B4M976 A0A0L0C211 A0A182SHE2 A0A182LRT3 B4P353 A0A1A9XRQ8 A0A182W0K3 A0A1A9W8I7 B3MV87 B4IEK1 A0A336LI50 A0A0L7RJM1 A0A0M8ZZD8 E2APR1 A0A2A3EF26 A0A088ADE6 A0A0B7AJV3 A0A026X5R4 A0A3L8DJ19 A0A0K8TTB2 A0A0B7AJI6 A0A154NW11 A0A1B6FH87 A0A0J7L395 A0A0P4WFF9 A0A158NVI5 E0VTK1 A0A2H4D133 A0A195BN03 A0A1B6ISW9 V4A538 F4WLF4 K7IVE7 A0A232EV28 A0A2J7RFU8 A0A3B3DW59 A0A067QVZ4 A0A1W4Y1D7 A0A2A2L411 A0A2H8TR00
A0A194Q971 A0A194RAI7 A0A0L7LBG6 A0A0T6AV65 D6WB96 A0A1Q3G293 Q17G46 A0A023EJW2 N6SY01 J3JX56 B4MWH2 A0A182TL42 A0A182XK69 A0A182KWY9 Q7QDJ9 A0A182HJR9 A0A182V8K1 A0A182K0J1 A0A2M3Z6P3 A0A1W4WZZ6 W5JPB3 A0A182IK21 T1E7U8 A0A182NKV0 A0A2M4BZ11 A0A084VZD2 A0A1I8M1A7 A0A1Y1MVF5 Q9VJZ6 A0A2M3Z6S5 A0A182R811 A0A2M4AQ69 A0A182QLC2 A0A2M4APD1 W8CDM7 A0A1L8ED64 A0A1L8DU36 V5H0Y1 A0A1I8Q318 A0A2M4APH6 A0A3B0KPJ4 B4JP71 A0A182PS52 A0A1B0D3Y8 B4GKA8 A0A034V9R6 Q29P85 A0A0K8WEH2 A0A1W4V8N9 U4U4Z2 B4KK55 B4Q4S2 A0A0M5J1Z2 B3NAY1 B4M976 A0A0L0C211 A0A182SHE2 A0A182LRT3 B4P353 A0A1A9XRQ8 A0A182W0K3 A0A1A9W8I7 B3MV87 B4IEK1 A0A336LI50 A0A0L7RJM1 A0A0M8ZZD8 E2APR1 A0A2A3EF26 A0A088ADE6 A0A0B7AJV3 A0A026X5R4 A0A3L8DJ19 A0A0K8TTB2 A0A0B7AJI6 A0A154NW11 A0A1B6FH87 A0A0J7L395 A0A0P4WFF9 A0A158NVI5 E0VTK1 A0A2H4D133 A0A195BN03 A0A1B6ISW9 V4A538 F4WLF4 K7IVE7 A0A232EV28 A0A2J7RFU8 A0A3B3DW59 A0A067QVZ4 A0A1W4Y1D7 A0A2A2L411 A0A2H8TR00
PDB
2YAN
E-value=2.91475e-35,
Score=369
Ontologies
GO
PANTHER
Topology
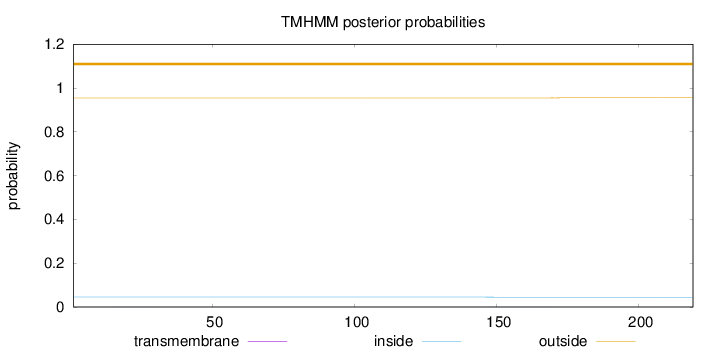
Length:
219
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01781
Exp number, first 60 AAs:
0.00303
Total prob of N-in:
0.04545
outside
1 - 219
Population Genetic Test Statistics
Pi
295.615629
Theta
196.764976
Tajima's D
1.804563
CLR
0.155355
CSRT
0.847607619619019
Interpretation
Uncertain
