Gene
KWMTBOMO03348 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006476
Annotation
cleavage_stimulation_factor_64-kDa_subunit_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.398
Sequence
CDS
ATGGATAAAAGTAAAGAAAAAGAGGAACAAAGCATCATGGATAAATCCATGAGATCTGTTTTCGTGGGTAATATACCATACGAAGCCACAGAAGAGAAATTAAAGGACATATTCAGTGAAGTAGGACCCGTCTTGTCTTTCAAACTTGTCTTCGATCGCGAAACCGGTAAACCCAAAGGTTATGGGTTTTGTGAATATAAAGACCAGGAAACGGCTTTGAGTGCCATGAGAAATCTCAATGGATACGAAATTGGTGGTCGTTCGTTAAGAGTAGATAATGCTTGTACAGAGAAATCGCGAATGGAAATGCAAGCTCTAATGCAAGGACCTCAGGTAGAAAATCCTTATGGTGATCCAGTAGAACCTGAAAAAGCACCTGAAGCAATCAGTAAAGCTGTGGCGACATTGCCTCCAGAACAGATGTTTGAACTGATGAAGCAAATGAAACTTTGTATTCAGAATAATCCTAATGAAGCTCGAAACATGCTTTTACAAAATCCGCAACTGGCATACGCGCTGCTACAGGCACAGGTCATAATGAGAATTGTCGATCCAGCCACTGCAGTTACAATGCTTCATCCAAGTAACCCTGTACCTCCAATCTTACTACCGGGTGATAAACCAGCCCAAACGAATACTTATATACCATCCAATCCACCACCAGCACAACCACCGATACACAATAATCCCGGACCGCCACCCAACCAGTACAACAATTCCCGTCCGTCCATGGATGTTGATCTTCGCGGGTCGCGAGCTCCCGGTCCCGCGGCGCCCCCTAACGCCATGCCCGCGCTGCTCGACCAGGACATGCGCAGTCTGCCGCCGGTCGCCCCGCACCCGGTACCGCCGCCGATGCATCATCCCCAAGATCCTAGTTACCCGAGAGATCCGCGGTTGGCCAATGCTCCACCGTACAACCCGGATCCGAGGGTGCGTTCAAGCGATCCACGAAACCAGCAGAAACCGCCACAGCAAATGCCTCCTGGCATGCCGAGCGCTGCACAGGCTAGGAACATTCAAGGTATTCCATCGGGTGCTTCTGATCAAGAGAAGGCCGCGTTGATAATGCAAGTGCTGCAGTTATCCGACGAACAGATCGCTTTGTTACCGCCCGAACAACGCGCCAGTATACTCCTGCTGAAAGAACAAATCGCGAAGAGCACACAGCAGCGTTAG
Protein
MDKSKEKEEQSIMDKSMRSVFVGNIPYEATEEKLKDIFSEVGPVLSFKLVFDRETGKPKGYGFCEYKDQETALSAMRNLNGYEIGGRSLRVDNACTEKSRMEMQALMQGPQVENPYGDPVEPEKAPEAISKAVATLPPEQMFELMKQMKLCIQNNPNEARNMLLQNPQLAYALLQAQVIMRIVDPATAVTMLHPSNPVPPILLPGDKPAQTNTYIPSNPPPAQPPIHNNPGPPPNQYNNSRPSMDVDLRGSRAPGPAAPPNAMPALLDQDMRSLPPVAPHPVPPPMHHPQDPSYPRDPRLANAPPYNPDPRVRSSDPRNQQKPPQQMPPGMPSAAQARNIQGIPSGASDQEKAALIMQVLQLSDEQIALLPPEQRASILLLKEQIAKSTQQR
Summary
Uniprot
EMBL
BABH01009113
BABH01009114
BABH01009115
BABH01009116
KQ460473
KPJ14517.1
+ More
ODYU01004378 SOQ44194.1 DQ497201 ABF55966.2 KQ971382 EEZ97191.2 CH478130 EAT33866.1 JXUM01087307 JXUM01117724 KQ566112 KQ563624 KXJ70390.1 KXJ73586.1 UFQT01000567 SSX25380.1 UFQS01000647 UFQT01000647 SSX05735.1 SSX26094.1 JRES01001362 JRES01000142 KNC23353.1 KNC33864.1 JXJN01014010 OUUW01000013 SPP87977.1 GANO01004064 JAB55807.1 GAPW01002101 JAC11497.1
ODYU01004378 SOQ44194.1 DQ497201 ABF55966.2 KQ971382 EEZ97191.2 CH478130 EAT33866.1 JXUM01087307 JXUM01117724 KQ566112 KQ563624 KXJ70390.1 KXJ73586.1 UFQT01000567 SSX25380.1 UFQS01000647 UFQT01000647 SSX05735.1 SSX26094.1 JRES01001362 JRES01000142 KNC23353.1 KNC33864.1 JXJN01014010 OUUW01000013 SPP87977.1 GANO01004064 JAB55807.1 GAPW01002101 JAC11497.1
Proteomes
Interpro
SUPFAM
SSF54928
SSF54928
Gene 3D
ProteinModelPortal
PDB
1P1T
E-value=3.63334e-38,
Score=397
Ontologies
GO
PANTHER
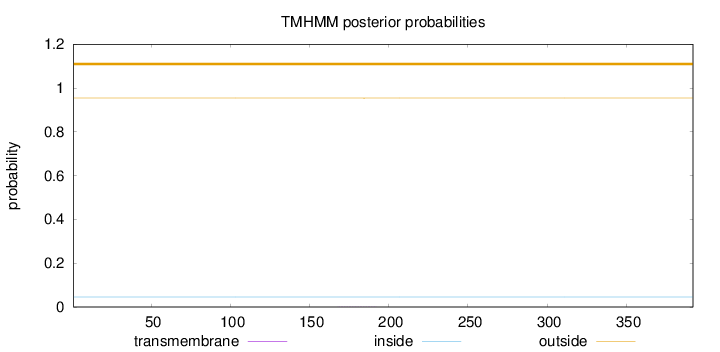
Topology
Length:
392
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01248
Exp number, first 60 AAs:
0.00015
Total prob of N-in:
0.04592
outside
1 - 392
Population Genetic Test Statistics
Pi
201.542476
Theta
165.883684
Tajima's D
0.586029
CLR
0.003406
CSRT
0.537073146342683
Interpretation
Uncertain
