Pre Gene Modal
BGIBMGA006475
Annotation
PREDICTED:_ras-related_C3_botulinum_toxin_substrate_1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.376 Nuclear Reliability : 1.603
Sequence
CDS
ATGTCTTCCGGAAGACCGATCAAGTGTGTGGTCGTTGGAGATGGTACCGTCGGAAAAACTTGTATGTTAATTTCATACACGACTGATAGCTTTCCAGGAGAATATGTGCCTACAGTTTTTGACAATTACTCGGCTCCAATGGTGGTGGATGGAGTGGCAGTATCTCTTGGTCTATGGGATACAGCGGGACAAGAAGACTACGACAGACTTAGACCTTTAAGCTATCCTCAAACTGATGTCTTCCTTATATGCTTCAGTGTAACAAGTCCCTCTTCATATGAAAATGTCACATCGAAATGGTACCCCGAAATAAAACATCACTGCCCCGATGCACCAATTATATTAGTGGGTACAAAAATCGATTTGAGAGATGATCGTGAAACGCTAAGTTTGCTGTCGGAACAAGGAATGTCTCCGCTGAAAAGGGAACAAGGACAGAAGCTCGCCAACAAAATTCGGGCCGTAAAATATATGGAGTGCTCAGCGCTAACACAAAGAGGCCTTAAACAGGTGTTCGACGAGGCTGTACGTGCAGTGTTAAGACCGGAACCACAAAAGAGACACCAGAGGAAGTGTTTGATTATGTAA
Protein
MSSGRPIKCVVVGDGTVGKTCMLISYTTDSFPGEYVPTVFDNYSAPMVVDGVAVSLGLWDTAGQEDYDRLRPLSYPQTDVFLICFSVTSPSSYENVTSKWYPEIKHHCPDAPIILVGTKIDLRDDRETLSLLSEQGMSPLKREQGQKLANKIRAVKYMECSALTQRGLKQVFDEAVRAVLRPEPQKRHQRKCLIM
Summary
Uniprot
A0A2A4JDN7
A0A194RAJ2
A0A2H1VTX6
S4P9B7
A0A212FI27
A0A1E1WK62
+ More
A0A194Q362 A0A1W4WPP0 D6W9A4 A0A1Y1MJ13 A0A336LIK7 A0A067QXA7 A0A1B6M7C1 A0A1B6FZ95 A0A0B4J2S0 W8AJG5 A0A1L8DRC1 A0A154PF17 A0A151J934 A0A195CN57 A0A0L7QPF8 F4WKU9 A0A195BKS3 V9ID57 A0A0B4J2P8 A0A151JVR7 E2BNN7 A0A158P3W9 A0A310SGQ3 E2AUD8 A0A151WV46 A0A026VYH3 A0A0A1WYL8 A0A1B0D022 A0A034VDP9 A0A0K8W8G4 A0A1Q3EYB3 B0X1Z9 Q1DH30 A0A182H083 A0A182YAN5 A0A0C9R0N8 U5EVN0 A0A182LRP6 A0A182IL89 A0A182T6T8 A0A023FAY4 A0A0A9VRS0 A0A182PGS2 A0A182U0U9 A0A182L9M2 A0A182WSE1 Q380U3 A0A182HL91 A0A182R656 A0A0L0BXV2 A0A1I8PY54 A0A1I8N252 A0A1I8PY86 A0A182V2L4 A0A182N421 A0A182Q4W9 A0A1B0G9P8 A0A1A9YD31 A0A1A9UD82 A0A1B0BUY6 A0A1A9Z7B9 A0A2M3ZDE2 A0A2M4AID3 W5JA36 A0A182F7L3 B4KB97 A0A0M3QXJ2 B4JRY0 R4G4I8 B4M627 A0A3B0JEE4 Q29BH0 B4GNY0 B4NAQ9 A0A1S3DLG0 A0A1J1HHY1 E0VQF2 T1JC27 A0A087T9E5 B4IGU2 Q7JWS8 B3LXI6 B4PRC0 B3P5Z4 A0A1W4UPN9 A0A224XXR2 A0A1D2NCH2 A0A1U6ZII7 A0A164TX58 A0A0P4WF52 A0A2A3E2M3 E9H2U3 A0A2H8TJY3 A0A0P6ID12
A0A194Q362 A0A1W4WPP0 D6W9A4 A0A1Y1MJ13 A0A336LIK7 A0A067QXA7 A0A1B6M7C1 A0A1B6FZ95 A0A0B4J2S0 W8AJG5 A0A1L8DRC1 A0A154PF17 A0A151J934 A0A195CN57 A0A0L7QPF8 F4WKU9 A0A195BKS3 V9ID57 A0A0B4J2P8 A0A151JVR7 E2BNN7 A0A158P3W9 A0A310SGQ3 E2AUD8 A0A151WV46 A0A026VYH3 A0A0A1WYL8 A0A1B0D022 A0A034VDP9 A0A0K8W8G4 A0A1Q3EYB3 B0X1Z9 Q1DH30 A0A182H083 A0A182YAN5 A0A0C9R0N8 U5EVN0 A0A182LRP6 A0A182IL89 A0A182T6T8 A0A023FAY4 A0A0A9VRS0 A0A182PGS2 A0A182U0U9 A0A182L9M2 A0A182WSE1 Q380U3 A0A182HL91 A0A182R656 A0A0L0BXV2 A0A1I8PY54 A0A1I8N252 A0A1I8PY86 A0A182V2L4 A0A182N421 A0A182Q4W9 A0A1B0G9P8 A0A1A9YD31 A0A1A9UD82 A0A1B0BUY6 A0A1A9Z7B9 A0A2M3ZDE2 A0A2M4AID3 W5JA36 A0A182F7L3 B4KB97 A0A0M3QXJ2 B4JRY0 R4G4I8 B4M627 A0A3B0JEE4 Q29BH0 B4GNY0 B4NAQ9 A0A1S3DLG0 A0A1J1HHY1 E0VQF2 T1JC27 A0A087T9E5 B4IGU2 Q7JWS8 B3LXI6 B4PRC0 B3P5Z4 A0A1W4UPN9 A0A224XXR2 A0A1D2NCH2 A0A1U6ZII7 A0A164TX58 A0A0P4WF52 A0A2A3E2M3 E9H2U3 A0A2H8TJY3 A0A0P6ID12
Pubmed
26354079
23622113
22118469
18362917
19820115
28004739
+ More
24845553 20075255 24495485 21719571 20798317 21347285 24508170 30249741 25830018 25348373 17510324 26483478 25244985 25474469 25401762 20966253 12364791 14747013 17210077 26108605 25315136 20920257 23761445 17994087 15632085 20566863 10847683 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 27289101 27385304 21292972
24845553 20075255 24495485 21719571 20798317 21347285 24508170 30249741 25830018 25348373 17510324 26483478 25244985 25474469 25401762 20966253 12364791 14747013 17210077 26108605 25315136 20920257 23761445 17994087 15632085 20566863 10847683 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 27289101 27385304 21292972
EMBL
NWSH01001882
PCG69808.1
KQ460473
KPJ14519.1
ODYU01004378
SOQ44196.1
+ More
GAIX01003814 JAA88746.1 AGBW02008443 OWR53392.1 GDQN01007920 GDQN01003736 JAT83134.1 JAT87318.1 KQ459551 KPI99976.1 KQ971312 EEZ99260.1 GEZM01030170 GEZM01030168 JAV85782.1 UFQS01000014 UFQT01000014 SSW97284.1 SSX17670.1 KK852854 KDR15004.1 GEBQ01008162 JAT31815.1 GECZ01014273 JAS55496.1 GAMC01020633 JAB85922.1 GFDF01005194 JAV08890.1 KQ434878 KZC09898.1 KQ979480 KYN21265.1 KQ977513 KYN02181.1 KQ414815 KOC60520.1 GL888206 EGI65191.1 KQ976453 KYM85325.1 JR040200 AEY59025.1 KQ981701 KYN37480.1 GL449438 EFN82685.1 ADTU01008608 KQ764027 OAD54630.1 GL442815 EFN62995.1 KQ982708 KYQ51802.1 KK107570 QOIP01000005 EZA48853.1 RLU23095.1 GBXI01010789 JAD03503.1 AJVK01021176 AJVK01021177 GAKP01018323 JAC40629.1 GDHF01011711 GDHF01005005 JAI40603.1 JAI47309.1 GFDL01014751 JAV20294.1 DS232275 EDS38929.1 CH899816 EAT32460.1 JXUM01022255 JXUM01022256 KQ560625 KXJ81541.1 GBYB01009704 JAG79471.1 GANO01001790 JAB58081.1 AXCM01005041 GBBI01000624 JAC18088.1 GBHO01045155 GBHO01034531 GBHO01021844 GBHO01021815 GBHO01016383 GBHO01016380 GBHO01016377 GBHO01013400 GBHO01011318 GBHO01007082 GBRD01003288 JAF98448.1 JAG09073.1 JAG21760.1 JAG21789.1 JAG27221.1 JAG27224.1 JAG27227.1 JAG30204.1 JAG32286.1 JAG36522.1 JAG62533.1 AAAB01008987 EAL38571.2 APCN01000885 JRES01001168 KNC24852.1 AXCN02000022 CCAG010002477 JXJN01020967 GGFM01005719 MBW26470.1 GGFK01007222 MBW40543.1 ADMH02001961 ETN60273.1 CH933806 EDW16825.1 CP012526 ALC45959.1 CH916373 EDV94520.1 GAHY01000736 JAA76774.1 CH940652 EDW59103.1 OUUW01000005 SPP80697.1 CM000070 EAL27028.1 CH479186 EDW38863.1 CH964232 EDW80873.1 CVRI01000003 CRK87020.1 DS235418 EEB15608.1 JH432049 KK114118 KFM61734.1 CH480837 CH684237 EDW49060.1 EDW56455.1 AF238044 AE014297 AY113279 AJ413188 AAF44665.1 AAF56727.1 AAF56728.1 AAM29284.1 AAN14120.1 CAC88352.1 CH902617 EDV43880.1 CM000160 EDW98484.1 CH954182 EDV53394.1 GFTR01003086 JAW13340.1 LJIJ01000092 ODN02931.1 KP765446 ALX18441.1 LRGB01001663 KZS10848.1 GDRN01035324 JAI67409.1 KZ288451 PBC25572.1 GL732587 EFX73948.1 GFXV01002662 MBW14467.1 GDIQ01006346 JAN88391.1
GAIX01003814 JAA88746.1 AGBW02008443 OWR53392.1 GDQN01007920 GDQN01003736 JAT83134.1 JAT87318.1 KQ459551 KPI99976.1 KQ971312 EEZ99260.1 GEZM01030170 GEZM01030168 JAV85782.1 UFQS01000014 UFQT01000014 SSW97284.1 SSX17670.1 KK852854 KDR15004.1 GEBQ01008162 JAT31815.1 GECZ01014273 JAS55496.1 GAMC01020633 JAB85922.1 GFDF01005194 JAV08890.1 KQ434878 KZC09898.1 KQ979480 KYN21265.1 KQ977513 KYN02181.1 KQ414815 KOC60520.1 GL888206 EGI65191.1 KQ976453 KYM85325.1 JR040200 AEY59025.1 KQ981701 KYN37480.1 GL449438 EFN82685.1 ADTU01008608 KQ764027 OAD54630.1 GL442815 EFN62995.1 KQ982708 KYQ51802.1 KK107570 QOIP01000005 EZA48853.1 RLU23095.1 GBXI01010789 JAD03503.1 AJVK01021176 AJVK01021177 GAKP01018323 JAC40629.1 GDHF01011711 GDHF01005005 JAI40603.1 JAI47309.1 GFDL01014751 JAV20294.1 DS232275 EDS38929.1 CH899816 EAT32460.1 JXUM01022255 JXUM01022256 KQ560625 KXJ81541.1 GBYB01009704 JAG79471.1 GANO01001790 JAB58081.1 AXCM01005041 GBBI01000624 JAC18088.1 GBHO01045155 GBHO01034531 GBHO01021844 GBHO01021815 GBHO01016383 GBHO01016380 GBHO01016377 GBHO01013400 GBHO01011318 GBHO01007082 GBRD01003288 JAF98448.1 JAG09073.1 JAG21760.1 JAG21789.1 JAG27221.1 JAG27224.1 JAG27227.1 JAG30204.1 JAG32286.1 JAG36522.1 JAG62533.1 AAAB01008987 EAL38571.2 APCN01000885 JRES01001168 KNC24852.1 AXCN02000022 CCAG010002477 JXJN01020967 GGFM01005719 MBW26470.1 GGFK01007222 MBW40543.1 ADMH02001961 ETN60273.1 CH933806 EDW16825.1 CP012526 ALC45959.1 CH916373 EDV94520.1 GAHY01000736 JAA76774.1 CH940652 EDW59103.1 OUUW01000005 SPP80697.1 CM000070 EAL27028.1 CH479186 EDW38863.1 CH964232 EDW80873.1 CVRI01000003 CRK87020.1 DS235418 EEB15608.1 JH432049 KK114118 KFM61734.1 CH480837 CH684237 EDW49060.1 EDW56455.1 AF238044 AE014297 AY113279 AJ413188 AAF44665.1 AAF56727.1 AAF56728.1 AAM29284.1 AAN14120.1 CAC88352.1 CH902617 EDV43880.1 CM000160 EDW98484.1 CH954182 EDV53394.1 GFTR01003086 JAW13340.1 LJIJ01000092 ODN02931.1 KP765446 ALX18441.1 LRGB01001663 KZS10848.1 GDRN01035324 JAI67409.1 KZ288451 PBC25572.1 GL732587 EFX73948.1 GFXV01002662 MBW14467.1 GDIQ01006346 JAN88391.1
Proteomes
UP000218220
UP000053240
UP000007151
UP000053268
UP000192223
UP000007266
+ More
UP000027135 UP000002358 UP000076502 UP000078492 UP000078542 UP000053825 UP000007755 UP000078540 UP000005203 UP000078541 UP000008237 UP000005205 UP000000311 UP000075809 UP000053097 UP000279307 UP000092462 UP000002320 UP000008820 UP000069940 UP000249989 UP000076408 UP000075883 UP000075880 UP000075901 UP000075885 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000075900 UP000037069 UP000095300 UP000095301 UP000075903 UP000075884 UP000075886 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000000673 UP000069272 UP000009192 UP000092553 UP000001070 UP000008792 UP000268350 UP000001819 UP000008744 UP000007798 UP000079169 UP000183832 UP000009046 UP000054359 UP000001292 UP000000803 UP000007801 UP000002282 UP000008711 UP000192221 UP000094527 UP000076858 UP000242457 UP000000305
UP000027135 UP000002358 UP000076502 UP000078492 UP000078542 UP000053825 UP000007755 UP000078540 UP000005203 UP000078541 UP000008237 UP000005205 UP000000311 UP000075809 UP000053097 UP000279307 UP000092462 UP000002320 UP000008820 UP000069940 UP000249989 UP000076408 UP000075883 UP000075880 UP000075901 UP000075885 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000075900 UP000037069 UP000095300 UP000095301 UP000075903 UP000075884 UP000075886 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000000673 UP000069272 UP000009192 UP000092553 UP000001070 UP000008792 UP000268350 UP000001819 UP000008744 UP000007798 UP000079169 UP000183832 UP000009046 UP000054359 UP000001292 UP000000803 UP000007801 UP000002282 UP000008711 UP000192221 UP000094527 UP000076858 UP000242457 UP000000305
PRIDE
Pfam
PF00071 Ras
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A2A4JDN7
A0A194RAJ2
A0A2H1VTX6
S4P9B7
A0A212FI27
A0A1E1WK62
+ More
A0A194Q362 A0A1W4WPP0 D6W9A4 A0A1Y1MJ13 A0A336LIK7 A0A067QXA7 A0A1B6M7C1 A0A1B6FZ95 A0A0B4J2S0 W8AJG5 A0A1L8DRC1 A0A154PF17 A0A151J934 A0A195CN57 A0A0L7QPF8 F4WKU9 A0A195BKS3 V9ID57 A0A0B4J2P8 A0A151JVR7 E2BNN7 A0A158P3W9 A0A310SGQ3 E2AUD8 A0A151WV46 A0A026VYH3 A0A0A1WYL8 A0A1B0D022 A0A034VDP9 A0A0K8W8G4 A0A1Q3EYB3 B0X1Z9 Q1DH30 A0A182H083 A0A182YAN5 A0A0C9R0N8 U5EVN0 A0A182LRP6 A0A182IL89 A0A182T6T8 A0A023FAY4 A0A0A9VRS0 A0A182PGS2 A0A182U0U9 A0A182L9M2 A0A182WSE1 Q380U3 A0A182HL91 A0A182R656 A0A0L0BXV2 A0A1I8PY54 A0A1I8N252 A0A1I8PY86 A0A182V2L4 A0A182N421 A0A182Q4W9 A0A1B0G9P8 A0A1A9YD31 A0A1A9UD82 A0A1B0BUY6 A0A1A9Z7B9 A0A2M3ZDE2 A0A2M4AID3 W5JA36 A0A182F7L3 B4KB97 A0A0M3QXJ2 B4JRY0 R4G4I8 B4M627 A0A3B0JEE4 Q29BH0 B4GNY0 B4NAQ9 A0A1S3DLG0 A0A1J1HHY1 E0VQF2 T1JC27 A0A087T9E5 B4IGU2 Q7JWS8 B3LXI6 B4PRC0 B3P5Z4 A0A1W4UPN9 A0A224XXR2 A0A1D2NCH2 A0A1U6ZII7 A0A164TX58 A0A0P4WF52 A0A2A3E2M3 E9H2U3 A0A2H8TJY3 A0A0P6ID12
A0A194Q362 A0A1W4WPP0 D6W9A4 A0A1Y1MJ13 A0A336LIK7 A0A067QXA7 A0A1B6M7C1 A0A1B6FZ95 A0A0B4J2S0 W8AJG5 A0A1L8DRC1 A0A154PF17 A0A151J934 A0A195CN57 A0A0L7QPF8 F4WKU9 A0A195BKS3 V9ID57 A0A0B4J2P8 A0A151JVR7 E2BNN7 A0A158P3W9 A0A310SGQ3 E2AUD8 A0A151WV46 A0A026VYH3 A0A0A1WYL8 A0A1B0D022 A0A034VDP9 A0A0K8W8G4 A0A1Q3EYB3 B0X1Z9 Q1DH30 A0A182H083 A0A182YAN5 A0A0C9R0N8 U5EVN0 A0A182LRP6 A0A182IL89 A0A182T6T8 A0A023FAY4 A0A0A9VRS0 A0A182PGS2 A0A182U0U9 A0A182L9M2 A0A182WSE1 Q380U3 A0A182HL91 A0A182R656 A0A0L0BXV2 A0A1I8PY54 A0A1I8N252 A0A1I8PY86 A0A182V2L4 A0A182N421 A0A182Q4W9 A0A1B0G9P8 A0A1A9YD31 A0A1A9UD82 A0A1B0BUY6 A0A1A9Z7B9 A0A2M3ZDE2 A0A2M4AID3 W5JA36 A0A182F7L3 B4KB97 A0A0M3QXJ2 B4JRY0 R4G4I8 B4M627 A0A3B0JEE4 Q29BH0 B4GNY0 B4NAQ9 A0A1S3DLG0 A0A1J1HHY1 E0VQF2 T1JC27 A0A087T9E5 B4IGU2 Q7JWS8 B3LXI6 B4PRC0 B3P5Z4 A0A1W4UPN9 A0A224XXR2 A0A1D2NCH2 A0A1U6ZII7 A0A164TX58 A0A0P4WF52 A0A2A3E2M3 E9H2U3 A0A2H8TJY3 A0A0P6ID12
PDB
5FI0
E-value=3.42788e-82,
Score=773
Ontologies
PATHWAY
GO
GO:0005525
GO:0007264
GO:0003924
GO:0019901
GO:0030036
GO:0005737
GO:0007266
GO:0043652
GO:0016477
GO:0007015
GO:0005886
GO:0000902
GO:0030838
GO:0051301
GO:0008258
GO:0008078
GO:0001737
GO:0031532
GO:0042067
GO:0007390
GO:0090303
GO:0007394
GO:0007254
GO:0007435
GO:0051017
GO:0035099
GO:0030032
GO:0004767
GO:0007391
GO:0002009
GO:0016021
GO:0051082
GO:0008270
GO:0033177
GO:0000287
GO:0003723
GO:0006270
GO:0042555
GO:0048384
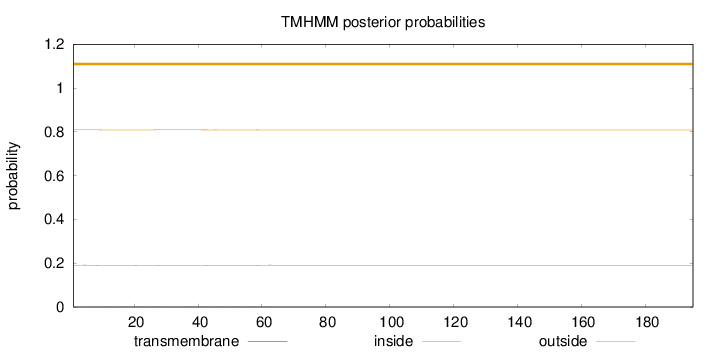
Topology
Length:
195
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04874
Exp number, first 60 AAs:
0.04581
Total prob of N-in:
0.19103
outside
1 - 195
Population Genetic Test Statistics
Pi
275.068931
Theta
180.741534
Tajima's D
1.679814
CLR
0.467168
CSRT
0.830608469576521
Interpretation
Uncertain
