Gene
KWMTBOMO03341
Pre Gene Modal
BGIBMGA014559
Annotation
PREDICTED:_peroxidase_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.279 Nuclear Reliability : 1.205
Sequence
CDS
ATGATCAAATTTCCACGTTTGACAATTGTTGTGATTGATAAACTTGTCGCCCGGCCAAAGGGTGACTGGAAACCTGAAGACTATGCGAGCGTTGGTGAACTCGTCCTCGACATTTCAATAAATCTGGCACGCAGATACGGTCTCTCGTACGAAGAGATTGAGAAAGGACTGCCTCTCATCGATACTTCCCGAACGCTTATCAGAGAGGTTTGCCCTCCTGTGTTCTCGCACGTGGAATGCCGGGCTGGCAAATACAGAAGATTTGACGGACTCTGCAATAACCTTAAGCATCCTACTTGGGGTTCCACGAATTCGCCTTTCCAGAGGTTAATTGGTCCCCTGTTCTCTGACGGCATTAACGCTCCTCGTATTTCACACACTGGTCATGATTTGCCGCTGTCGCGTGTGGTGTCGCGCACCATGCATCCGGACGACGGTTTCCACGACCACGCGGGCACCGTAATGGTCATAGCCTGGGGACAGTTCATGGACCACGATTATACACTAACCGGAACTCCATTAGGTAACAATAACAATATACATTTCGTGGTGGGTGTCATGTTTTGA
Protein
MIKFPRLTIVVIDKLVARPKGDWKPEDYASVGELVLDISINLARRYGLSYEEIEKGLPLIDTSRTLIREVCPPVFSHVECRAGKYRRFDGLCNNLKHPTWGSTNSPFQRLIGPLFSDGINAPRISHTGHDLPLSRVVSRTMHPDDGFHDHAGTVMVIAWGQFMDHDYTLTGTPLGNNNNIHFVVGVMF
Summary
Uniprot
I4DR22
A0A194RA74
A0A194Q4P1
A0A2A4JCW1
B4IBH4
A0A1B0CP07
+ More
B3P3D6 Q9VEJ9 B4QVQ3 B4PVB5 A0A1W4V5L9 B4N9U3 A0A0M4ENK1 B3LVQ7 B4JTV2 B4LY40 A0A3B0JIT3 B4K7M3 Q29A85 A0A3B0JK03 B4G2M3 W8B8J8 A0A1I8PYQ5 A0A139WB90 A0A034VY50 A0A1I8MNE2 A0A0A1WQV2 A0A0K8VHI5 A0A0L0BW90 A0A182L691 A0A026VUC4 H9JYD9 A0A084WPT6 A0A182UD07 N6TF03 A0A182J7R2 A0A182RER6 A0A182G2M9 A0A182X7C0 A0A182V3N3 A0A182HIR6 A0A2M4BF79 Q7PFV3 A0A182VVM1 A0A182MQ32 A0A2M4BER8 A0A182Q7S7 A0A182K6T6 Q17IJ2 A0A182YCK3 A0A336M8J4 A0A182PK17 W5JN90 E9J8W4 A0A1J1I9G2 A0A1W4WW68 A0A182N0A1 B0XF69 A0A158NRH1 A0A195BD13 U4UXW9 A0A1B6KKX5 E0W1X9 A0A0J7L2M7 A0A195FU35 A0A310SHR8 A0A1B6GGR6 A0A1B6FH99 E2AXD6 A0A0A9Z5Q6 A0A195C4S7 A0A026WBZ6 A0A195DI97 A0A023F4G3 A0A3L8DGI6 A0A2R7WUG5 A0A2P8XJW1 A0A1B6CLE7 A0A0L7QJS1 A0A088ADW6 T1HLZ0 A0A146MBA6 F4WBP1 A0A2A3E3H2 A0A2J7QI05 A0A232F1P3 A0A154PF84 K7IQN3 A0A2S2QQ56 A0A0C9QE97 A0A0C9RV54 J9K3L8 A0A1A9WNS0 A0A1B0A7L8 A0A1A9X627 A0A2S2Q2H2
B3P3D6 Q9VEJ9 B4QVQ3 B4PVB5 A0A1W4V5L9 B4N9U3 A0A0M4ENK1 B3LVQ7 B4JTV2 B4LY40 A0A3B0JIT3 B4K7M3 Q29A85 A0A3B0JK03 B4G2M3 W8B8J8 A0A1I8PYQ5 A0A139WB90 A0A034VY50 A0A1I8MNE2 A0A0A1WQV2 A0A0K8VHI5 A0A0L0BW90 A0A182L691 A0A026VUC4 H9JYD9 A0A084WPT6 A0A182UD07 N6TF03 A0A182J7R2 A0A182RER6 A0A182G2M9 A0A182X7C0 A0A182V3N3 A0A182HIR6 A0A2M4BF79 Q7PFV3 A0A182VVM1 A0A182MQ32 A0A2M4BER8 A0A182Q7S7 A0A182K6T6 Q17IJ2 A0A182YCK3 A0A336M8J4 A0A182PK17 W5JN90 E9J8W4 A0A1J1I9G2 A0A1W4WW68 A0A182N0A1 B0XF69 A0A158NRH1 A0A195BD13 U4UXW9 A0A1B6KKX5 E0W1X9 A0A0J7L2M7 A0A195FU35 A0A310SHR8 A0A1B6GGR6 A0A1B6FH99 E2AXD6 A0A0A9Z5Q6 A0A195C4S7 A0A026WBZ6 A0A195DI97 A0A023F4G3 A0A3L8DGI6 A0A2R7WUG5 A0A2P8XJW1 A0A1B6CLE7 A0A0L7QJS1 A0A088ADW6 T1HLZ0 A0A146MBA6 F4WBP1 A0A2A3E3H2 A0A2J7QI05 A0A232F1P3 A0A154PF84 K7IQN3 A0A2S2QQ56 A0A0C9QE97 A0A0C9RV54 J9K3L8 A0A1A9WNS0 A0A1B0A7L8 A0A1A9X627 A0A2S2Q2H2
Pubmed
22651552
26354079
17994087
10731132
12537568
12537572
+ More
12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 24495485 18362917 19820115 25348373 25315136 25830018 26108605 20966253 24508170 19121390 24438588 23537049 26483478 12364791 17510324 25244985 20920257 23761445 21282665 21347285 20566863 20798317 25401762 25474469 30249741 29403074 26823975 21719571 28648823 20075255
12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 24495485 18362917 19820115 25348373 25315136 25830018 26108605 20966253 24508170 19121390 24438588 23537049 26483478 12364791 17510324 25244985 20920257 23761445 21282665 21347285 20566863 20798317 25401762 25474469 30249741 29403074 26823975 21719571 28648823 20075255
EMBL
AK404900
BAM20362.1
KQ460473
KPJ14522.1
KQ459551
KPI99979.1
+ More
NWSH01001882 PCG69801.1 CH480827 EDW44732.1 AJWK01021274 CH954181 EDV48716.1 AE014297 BT021336 AAF55422.2 AAX33484.1 CM000364 EDX12566.1 CM000160 EDW95784.1 CH964232 EDW80658.2 CP012526 ALC46288.1 CH902617 EDV43681.1 CH916374 EDV91531.1 CH940650 EDW67928.2 OUUW01000005 SPP80653.1 CH933806 EDW15367.1 CM000070 EAL27465.3 SPP80652.1 CH479179 EDW24068.1 GAMC01017079 GAMC01017078 JAB89476.1 KQ971374 KYB25155.1 GAKP01011588 JAC47364.1 GBXI01013256 JAD01036.1 GDHF01013996 JAI38318.1 JRES01001246 KNC24292.1 KK111212 EZA46454.1 BABH01043176 ATLV01025130 KE525369 KFB52230.1 APGK01040545 APGK01040546 APGK01040547 KB740983 ENN76348.1 JXUM01139699 KQ568940 KXJ68831.1 APCN01004052 APCN01004053 GGFJ01002470 MBW51611.1 AAAB01008846 EAA45172.5 AXCM01014078 GGFJ01002404 MBW51545.1 AXCN02001245 CH477239 EAT46477.2 UFQT01000247 SSX22358.1 ADMH02001040 ETN64355.1 GL769162 EFZ10729.1 CVRI01000044 CRK96843.1 DS232903 EDS26535.1 ADTU01024028 ADTU01024029 ADTU01024030 ADTU01024031 KQ976511 KYM82461.1 KB632410 ERL95211.1 GEBQ01027878 JAT12099.1 DS235873 EEB19573.1 LBMM01001090 KMQ96941.1 KQ981280 KYN43399.1 KQ762935 OAD55249.1 GECZ01008131 JAS61638.1 GECZ01020214 JAS49555.1 GL443548 EFN61916.1 GBHO01003775 JAG39829.1 KQ978317 KYM95181.1 KK107279 EZA53580.1 KQ980824 KYN12613.1 GBBI01002306 JAC16406.1 QOIP01000008 RLU19431.1 KK855584 PTY23193.1 PYGN01001895 PSN32287.1 GEDC01023001 JAS14297.1 KQ415080 KOC58825.1 ACPB03014772 GDHC01002532 JAQ16097.1 GL888066 EGI68319.1 KZ288416 PBC26034.1 NEVH01013964 PNF28220.1 NNAY01001240 OXU24635.1 KQ434878 KZC09890.1 GGMS01010654 MBY79857.1 GBYB01012733 JAG82500.1 GBYB01012735 JAG82502.1 ABLF02030854 ABLF02030857 GGMS01002755 MBY71958.1
NWSH01001882 PCG69801.1 CH480827 EDW44732.1 AJWK01021274 CH954181 EDV48716.1 AE014297 BT021336 AAF55422.2 AAX33484.1 CM000364 EDX12566.1 CM000160 EDW95784.1 CH964232 EDW80658.2 CP012526 ALC46288.1 CH902617 EDV43681.1 CH916374 EDV91531.1 CH940650 EDW67928.2 OUUW01000005 SPP80653.1 CH933806 EDW15367.1 CM000070 EAL27465.3 SPP80652.1 CH479179 EDW24068.1 GAMC01017079 GAMC01017078 JAB89476.1 KQ971374 KYB25155.1 GAKP01011588 JAC47364.1 GBXI01013256 JAD01036.1 GDHF01013996 JAI38318.1 JRES01001246 KNC24292.1 KK111212 EZA46454.1 BABH01043176 ATLV01025130 KE525369 KFB52230.1 APGK01040545 APGK01040546 APGK01040547 KB740983 ENN76348.1 JXUM01139699 KQ568940 KXJ68831.1 APCN01004052 APCN01004053 GGFJ01002470 MBW51611.1 AAAB01008846 EAA45172.5 AXCM01014078 GGFJ01002404 MBW51545.1 AXCN02001245 CH477239 EAT46477.2 UFQT01000247 SSX22358.1 ADMH02001040 ETN64355.1 GL769162 EFZ10729.1 CVRI01000044 CRK96843.1 DS232903 EDS26535.1 ADTU01024028 ADTU01024029 ADTU01024030 ADTU01024031 KQ976511 KYM82461.1 KB632410 ERL95211.1 GEBQ01027878 JAT12099.1 DS235873 EEB19573.1 LBMM01001090 KMQ96941.1 KQ981280 KYN43399.1 KQ762935 OAD55249.1 GECZ01008131 JAS61638.1 GECZ01020214 JAS49555.1 GL443548 EFN61916.1 GBHO01003775 JAG39829.1 KQ978317 KYM95181.1 KK107279 EZA53580.1 KQ980824 KYN12613.1 GBBI01002306 JAC16406.1 QOIP01000008 RLU19431.1 KK855584 PTY23193.1 PYGN01001895 PSN32287.1 GEDC01023001 JAS14297.1 KQ415080 KOC58825.1 ACPB03014772 GDHC01002532 JAQ16097.1 GL888066 EGI68319.1 KZ288416 PBC26034.1 NEVH01013964 PNF28220.1 NNAY01001240 OXU24635.1 KQ434878 KZC09890.1 GGMS01010654 MBY79857.1 GBYB01012733 JAG82500.1 GBYB01012735 JAG82502.1 ABLF02030854 ABLF02030857 GGMS01002755 MBY71958.1
Proteomes
UP000053240
UP000053268
UP000218220
UP000001292
UP000092461
UP000008711
+ More
UP000000803 UP000000304 UP000002282 UP000192221 UP000007798 UP000092553 UP000007801 UP000001070 UP000008792 UP000268350 UP000009192 UP000001819 UP000008744 UP000095300 UP000007266 UP000095301 UP000037069 UP000075882 UP000053097 UP000005204 UP000030765 UP000075902 UP000019118 UP000075880 UP000075900 UP000069940 UP000249989 UP000076407 UP000075903 UP000075840 UP000007062 UP000075920 UP000075883 UP000075886 UP000075881 UP000008820 UP000076408 UP000075885 UP000000673 UP000183832 UP000192223 UP000075884 UP000002320 UP000005205 UP000078540 UP000030742 UP000009046 UP000036403 UP000078541 UP000000311 UP000078542 UP000078492 UP000279307 UP000245037 UP000053825 UP000005203 UP000015103 UP000007755 UP000242457 UP000235965 UP000215335 UP000076502 UP000002358 UP000007819 UP000091820 UP000092445 UP000092443
UP000000803 UP000000304 UP000002282 UP000192221 UP000007798 UP000092553 UP000007801 UP000001070 UP000008792 UP000268350 UP000009192 UP000001819 UP000008744 UP000095300 UP000007266 UP000095301 UP000037069 UP000075882 UP000053097 UP000005204 UP000030765 UP000075902 UP000019118 UP000075880 UP000075900 UP000069940 UP000249989 UP000076407 UP000075903 UP000075840 UP000007062 UP000075920 UP000075883 UP000075886 UP000075881 UP000008820 UP000076408 UP000075885 UP000000673 UP000183832 UP000192223 UP000075884 UP000002320 UP000005205 UP000078540 UP000030742 UP000009046 UP000036403 UP000078541 UP000000311 UP000078542 UP000078492 UP000279307 UP000245037 UP000053825 UP000005203 UP000015103 UP000007755 UP000242457 UP000235965 UP000215335 UP000076502 UP000002358 UP000007819 UP000091820 UP000092445 UP000092443
Pfam
PF03098 An_peroxidase
Interpro
SUPFAM
SSF48113
SSF48113
Gene 3D
ProteinModelPortal
I4DR22
A0A194RA74
A0A194Q4P1
A0A2A4JCW1
B4IBH4
A0A1B0CP07
+ More
B3P3D6 Q9VEJ9 B4QVQ3 B4PVB5 A0A1W4V5L9 B4N9U3 A0A0M4ENK1 B3LVQ7 B4JTV2 B4LY40 A0A3B0JIT3 B4K7M3 Q29A85 A0A3B0JK03 B4G2M3 W8B8J8 A0A1I8PYQ5 A0A139WB90 A0A034VY50 A0A1I8MNE2 A0A0A1WQV2 A0A0K8VHI5 A0A0L0BW90 A0A182L691 A0A026VUC4 H9JYD9 A0A084WPT6 A0A182UD07 N6TF03 A0A182J7R2 A0A182RER6 A0A182G2M9 A0A182X7C0 A0A182V3N3 A0A182HIR6 A0A2M4BF79 Q7PFV3 A0A182VVM1 A0A182MQ32 A0A2M4BER8 A0A182Q7S7 A0A182K6T6 Q17IJ2 A0A182YCK3 A0A336M8J4 A0A182PK17 W5JN90 E9J8W4 A0A1J1I9G2 A0A1W4WW68 A0A182N0A1 B0XF69 A0A158NRH1 A0A195BD13 U4UXW9 A0A1B6KKX5 E0W1X9 A0A0J7L2M7 A0A195FU35 A0A310SHR8 A0A1B6GGR6 A0A1B6FH99 E2AXD6 A0A0A9Z5Q6 A0A195C4S7 A0A026WBZ6 A0A195DI97 A0A023F4G3 A0A3L8DGI6 A0A2R7WUG5 A0A2P8XJW1 A0A1B6CLE7 A0A0L7QJS1 A0A088ADW6 T1HLZ0 A0A146MBA6 F4WBP1 A0A2A3E3H2 A0A2J7QI05 A0A232F1P3 A0A154PF84 K7IQN3 A0A2S2QQ56 A0A0C9QE97 A0A0C9RV54 J9K3L8 A0A1A9WNS0 A0A1B0A7L8 A0A1A9X627 A0A2S2Q2H2
B3P3D6 Q9VEJ9 B4QVQ3 B4PVB5 A0A1W4V5L9 B4N9U3 A0A0M4ENK1 B3LVQ7 B4JTV2 B4LY40 A0A3B0JIT3 B4K7M3 Q29A85 A0A3B0JK03 B4G2M3 W8B8J8 A0A1I8PYQ5 A0A139WB90 A0A034VY50 A0A1I8MNE2 A0A0A1WQV2 A0A0K8VHI5 A0A0L0BW90 A0A182L691 A0A026VUC4 H9JYD9 A0A084WPT6 A0A182UD07 N6TF03 A0A182J7R2 A0A182RER6 A0A182G2M9 A0A182X7C0 A0A182V3N3 A0A182HIR6 A0A2M4BF79 Q7PFV3 A0A182VVM1 A0A182MQ32 A0A2M4BER8 A0A182Q7S7 A0A182K6T6 Q17IJ2 A0A182YCK3 A0A336M8J4 A0A182PK17 W5JN90 E9J8W4 A0A1J1I9G2 A0A1W4WW68 A0A182N0A1 B0XF69 A0A158NRH1 A0A195BD13 U4UXW9 A0A1B6KKX5 E0W1X9 A0A0J7L2M7 A0A195FU35 A0A310SHR8 A0A1B6GGR6 A0A1B6FH99 E2AXD6 A0A0A9Z5Q6 A0A195C4S7 A0A026WBZ6 A0A195DI97 A0A023F4G3 A0A3L8DGI6 A0A2R7WUG5 A0A2P8XJW1 A0A1B6CLE7 A0A0L7QJS1 A0A088ADW6 T1HLZ0 A0A146MBA6 F4WBP1 A0A2A3E3H2 A0A2J7QI05 A0A232F1P3 A0A154PF84 K7IQN3 A0A2S2QQ56 A0A0C9QE97 A0A0C9RV54 J9K3L8 A0A1A9WNS0 A0A1B0A7L8 A0A1A9X627 A0A2S2Q2H2
PDB
5MFA
E-value=4.87566e-10,
Score=150
Ontologies
GO
Topology
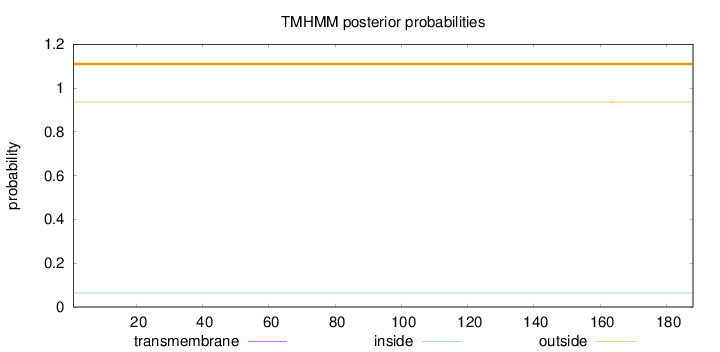
Length:
188
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01009
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.06421
outside
1 - 188
Population Genetic Test Statistics
Pi
279.668721
Theta
193.603859
Tajima's D
1.298002
CLR
0.045108
CSRT
0.736713164341783
Interpretation
Uncertain
