Pre Gene Modal
BGIBMGA006473
Annotation
PREDICTED:_mannose-6-phosphate_isomerase_[Amyelois_transitella]
Full name
Mannose-6-phosphate isomerase
Alternative Name
Phosphohexomutase
Phosphomannose isomerase
Phosphomannose isomerase
Location in the cell
Cytoplasmic Reliability : 1.949
Sequence
CDS
ATGGAATTGCAGTGCAAAGCTTTGCATTATGATTGGGGCAAAGTTGGCTCTGAGAGTATAGTTGCTAGGCTGTTACATAGTGCTGATTCTAGAATTGACATAGATCAAACAAAACCTTATGCTGAGTTATGGATGGGAACTCATCCTAATGGTCCTTCTCTAATTATTGAGAGAAATATTTTATTGTCTGACTACCTCAAAGAAAATCTTGATGCCATTGGGCCAGCTGTAAGGAAAAAATTTGGTGTAGCCATACCTTTCCTTTTGAAGATATTGTCTATCAAAAAGGCCCTGTCTATACAAGCCCATCCCAACAAGGAACATGCTGAACAGCTACACAAAAACTTCCCTGATATGTACAAAGATCCAAATCACAAGCCCGAGATTGCCATTGCCTTGACATCATTTGAGGCTCTTTGTGGATTTAGGCCTTTGTCTGAAATCAGGGAGTTCTTAAAAAAACTTCCTGAACTTACCAACATTCTATCCAAAGAAGCTGTTCACAGAGTGCTCACTAATCAGGATCAAGGAGACAATGACAAAATTCTAAGGCCATTGTTTGAGTCACTGATGACTTGTGAAAAAGCTTCTGTTTCCAACAGCTTGCAAAACTTGATCAACAGGCTTGAGAAAGATGATGCATCAAATCAAACATTTTTACTCTATTCTCTTCTGCGTCGCTTACATAGTGATTTCCCTGGAGATGTTGGCTGTTGGGGTCCATACTTCTTGAATTATATTGTGCTGCACCCAGGCCAAGCTATTTTCTTGAAGCCGAACCTGCCACACGCCTACTTGAGTGGAGATTGTGTAGAATGCATGGCATGTTCTGATAATGTGATCCGTGCTGGCTTGACTCCGAAACATATTGATGTGCCCACACTAGTCGATATGCTGGACTATAATAGCTATGGAAAAGATCAACTCCTATTCCACCCCCAATTAGAAGATGCAAATAGTTGTATTTGGCGTCCTCCGGTGCCTGATTTTGCTGTTGTGAAGATAAATGTGGTGGGTCCGGAGCCGTACAACACCCAGATCCGGCCCAGTCCGAGTCTCCTCATAGTGACGTCAGGGGCGGGCTCGGCGTGCGACACGGAGCCGGTGCCCGTCCGTGCCGGGGTCGTGCTGTTCCTGAAGGCGAGCCGTCAGCTCACACTCACGCCCACAGCTGACGGGCACATCGAGGCCTACCAGGCCATATGCAACGTATGA
Protein
MELQCKALHYDWGKVGSESIVARLLHSADSRIDIDQTKPYAELWMGTHPNGPSLIIERNILLSDYLKENLDAIGPAVRKKFGVAIPFLLKILSIKKALSIQAHPNKEHAEQLHKNFPDMYKDPNHKPEIAIALTSFEALCGFRPLSEIREFLKKLPELTNILSKEAVHRVLTNQDQGDNDKILRPLFESLMTCEKASVSNSLQNLINRLEKDDASNQTFLLYSLLRRLHSDFPGDVGCWGPYFLNYIVLHPGQAIFLKPNLPHAYLSGDCVECMACSDNVIRAGLTPKHIDVPTLVDMLDYNSYGKDQLLFHPQLEDANSCIWRPPVPDFAVVKINVVGPEPYNTQIRPSPSLLIVTSGAGSACDTEPVPVRAGVVLFLKASRQLTLTPTADGHIEAYQAICNV
Summary
Description
Involved in the synthesis of the GDP-mannose and dolichol-phosphate-mannose required for a number of critical mannosyl transfer reactions.
Catalytic Activity
D-mannose 6-phosphate = D-fructose 6-phosphate
Cofactor
Zn(2+)
Similarity
Belongs to the mannose-6-phosphate isomerase type 1 family.
Keywords
Acetylation
Alternative splicing
Complete proteome
Congenital disorder of glycosylation
Cytoplasm
Direct protein sequencing
Disease mutation
Isomerase
Metal-binding
Phosphoprotein
Reference proteome
Zinc
Feature
chain Mannose-6-phosphate isomerase
splice variant In isoform 2.
sequence variant In CDG1B; dbSNP:rs764835081.
splice variant In isoform 2.
sequence variant In CDG1B; dbSNP:rs764835081.
Uniprot
H9JAC8
A0A2A4JHC2
A0A2H1VVA5
A0A194R9L3
A0A194Q981
A0A212F4K2
+ More
A0A2A3E394 A0A088AHA0 A0A0M9A0D6 A0A0L7QST5 A0A154PCE3 A0A1B6FT62 A0A1B6MFM9 E2ARN2 A0A1B6EHC0 U4U4R5 A0A158NAH7 A0A151X534 D1ZZV1 A0A067QXA2 A0A0C9RX77 A0A151I5Q0 A0A2J7QHS1 A0A1Y1JXN4 C3ZF62 A0A026WK70 A0A195E4Y5 F6YAT8 A0A195EWE4 A0A164RYL5 A0A0P5I5N2 F4X239 A0A0P5QDA1 A0A0P6JWG5 A0A0P5PEQ5 A0A0P4XL22 E9GEW1 A0A2J7QHS8 A0A0P6BXW0 A0A0P5WY92 A0A0P5P0F0 A0A310SE35 G1SUK4 E9I9L0 A0A0P5P741 J3JX02 A0A2Y9DHN1 A0A2Y9HJA3 A0A2U3YUA8 A0A0P5ZL69 K7J6J4 V9KPF4 A0A0D9RPC6 G5AL67 U3CWX3 A0A384DIR6 A0A384DIV2 U3DUA7 A0A096NMS2 H2NNT1 H0VHT6 A0A2K5YTF3 G2HIF8 G3RFM0 P34949 H2Q9T4 A5A6K3 A0A2K5JTJ0 A0A2K5S6P1 A0A1A6H3X0 A0A1D5Q678 Q8HXX2 A0A2Y9K7X3
A0A2A3E394 A0A088AHA0 A0A0M9A0D6 A0A0L7QST5 A0A154PCE3 A0A1B6FT62 A0A1B6MFM9 E2ARN2 A0A1B6EHC0 U4U4R5 A0A158NAH7 A0A151X534 D1ZZV1 A0A067QXA2 A0A0C9RX77 A0A151I5Q0 A0A2J7QHS1 A0A1Y1JXN4 C3ZF62 A0A026WK70 A0A195E4Y5 F6YAT8 A0A195EWE4 A0A164RYL5 A0A0P5I5N2 F4X239 A0A0P5QDA1 A0A0P6JWG5 A0A0P5PEQ5 A0A0P4XL22 E9GEW1 A0A2J7QHS8 A0A0P6BXW0 A0A0P5WY92 A0A0P5P0F0 A0A310SE35 G1SUK4 E9I9L0 A0A0P5P741 J3JX02 A0A2Y9DHN1 A0A2Y9HJA3 A0A2U3YUA8 A0A0P5ZL69 K7J6J4 V9KPF4 A0A0D9RPC6 G5AL67 U3CWX3 A0A384DIR6 A0A384DIV2 U3DUA7 A0A096NMS2 H2NNT1 H0VHT6 A0A2K5YTF3 G2HIF8 G3RFM0 P34949 H2Q9T4 A5A6K3 A0A2K5JTJ0 A0A2K5S6P1 A0A1A6H3X0 A0A1D5Q678 Q8HXX2 A0A2Y9K7X3
EC Number
5.3.1.8
Pubmed
19121390
26354079
22118469
20798317
23537049
21347285
+ More
18362917 19820115 24845553 28004739 18563158 24508170 18464734 21719571 21292972 21993624 21282665 22516182 20075255 24402279 21993625 25243066 21484476 22398555 8307007 10980531 14702039 15489334 19413330 21269460 24275569 9585601 9525984 11134235 11350186 12357336 12414827 16136131 17574350 17431167
18362917 19820115 24845553 28004739 18563158 24508170 18464734 21719571 21292972 21993624 21282665 22516182 20075255 24402279 21993625 25243066 21484476 22398555 8307007 10980531 14702039 15489334 19413330 21269460 24275569 9585601 9525984 11134235 11350186 12357336 12414827 16136131 17574350 17431167
EMBL
BABH01009101
NWSH01001571
PCG70830.1
ODYU01004646
SOQ44748.1
KQ460473
+ More
KPJ14523.1 KQ459551 KPI99980.1 AGBW02010347 OWR48675.1 KZ288447 PBC25609.1 KQ435779 KOX74805.1 KQ414755 KOC61697.1 KQ434870 KZC09457.1 GECZ01016389 JAS53380.1 GEBQ01011757 GEBQ01005227 JAT28220.1 JAT34750.1 GL442157 EFN63894.1 GEDC01000024 JAS37274.1 KB631724 ERL85596.1 ADTU01010310 ADTU01010311 KQ982519 KYQ55525.1 KQ971338 EFA01794.1 KK852854 KDR14999.1 GBYB01013697 GBYB01013699 GBYB01013700 JAG83464.1 JAG83466.1 JAG83467.1 KQ976413 KYM90424.1 NEVH01013964 PNF28131.1 GEZM01102797 JAV51786.1 GG666612 EEN49368.1 KK107167 EZA56447.1 KQ979609 KYN20228.1 KQ981954 KYN32212.1 LRGB01002121 KZS09065.1 GDIQ01218458 JAK33267.1 GL888565 EGI59455.1 GDIQ01125302 JAL26424.1 GDIQ01005881 JAN88856.1 GDIQ01129640 JAL22086.1 GDIP01241094 JAI82307.1 GL732541 EFX81906.1 PNF28129.1 GDIP01023552 JAM80163.1 GDIP01079849 JAM23866.1 GDIQ01137039 JAL14687.1 KQ760792 OAD59000.1 GL761846 EFZ22736.1 GDIQ01137038 JAL14688.1 BT127770 AEE62732.1 GDIP01042804 JAM60911.1 AAZX01001318 JW868027 AFP00545.1 AQIB01139401 AQIB01139402 JH165769 GEBF01003856 EHA97777.1 JAN99776.1 GAMT01006170 GAMR01010003 GAMQ01007737 JAB05691.1 JAB23929.1 JAB34114.1 GAMS01009781 GAMP01009037 JAB13355.1 JAB43718.1 AHZZ02031288 ABGA01340157 ABGA01340158 ABGA01340159 NDHI03003463 PNJ43296.1 AAKN02050770 AK306522 GABC01005678 GABF01010277 GABD01005181 GABE01009275 NBAG03000345 BAK63516.1 JAA05660.1 JAA11868.1 JAA27919.1 JAA35464.1 PNI37779.1 CABD030097140 X76057 AF227218 AF227216 AF227217 AK292374 CH471136 BC017351 BC046357 AACZ04060366 AB222131 LZPO01055098 OBS72520.1 JSUE03038719 JSUE03038720 JSUE03038721 AB083318
KPJ14523.1 KQ459551 KPI99980.1 AGBW02010347 OWR48675.1 KZ288447 PBC25609.1 KQ435779 KOX74805.1 KQ414755 KOC61697.1 KQ434870 KZC09457.1 GECZ01016389 JAS53380.1 GEBQ01011757 GEBQ01005227 JAT28220.1 JAT34750.1 GL442157 EFN63894.1 GEDC01000024 JAS37274.1 KB631724 ERL85596.1 ADTU01010310 ADTU01010311 KQ982519 KYQ55525.1 KQ971338 EFA01794.1 KK852854 KDR14999.1 GBYB01013697 GBYB01013699 GBYB01013700 JAG83464.1 JAG83466.1 JAG83467.1 KQ976413 KYM90424.1 NEVH01013964 PNF28131.1 GEZM01102797 JAV51786.1 GG666612 EEN49368.1 KK107167 EZA56447.1 KQ979609 KYN20228.1 KQ981954 KYN32212.1 LRGB01002121 KZS09065.1 GDIQ01218458 JAK33267.1 GL888565 EGI59455.1 GDIQ01125302 JAL26424.1 GDIQ01005881 JAN88856.1 GDIQ01129640 JAL22086.1 GDIP01241094 JAI82307.1 GL732541 EFX81906.1 PNF28129.1 GDIP01023552 JAM80163.1 GDIP01079849 JAM23866.1 GDIQ01137039 JAL14687.1 KQ760792 OAD59000.1 GL761846 EFZ22736.1 GDIQ01137038 JAL14688.1 BT127770 AEE62732.1 GDIP01042804 JAM60911.1 AAZX01001318 JW868027 AFP00545.1 AQIB01139401 AQIB01139402 JH165769 GEBF01003856 EHA97777.1 JAN99776.1 GAMT01006170 GAMR01010003 GAMQ01007737 JAB05691.1 JAB23929.1 JAB34114.1 GAMS01009781 GAMP01009037 JAB13355.1 JAB43718.1 AHZZ02031288 ABGA01340157 ABGA01340158 ABGA01340159 NDHI03003463 PNJ43296.1 AAKN02050770 AK306522 GABC01005678 GABF01010277 GABD01005181 GABE01009275 NBAG03000345 BAK63516.1 JAA05660.1 JAA11868.1 JAA27919.1 JAA35464.1 PNI37779.1 CABD030097140 X76057 AF227218 AF227216 AF227217 AK292374 CH471136 BC017351 BC046357 AACZ04060366 AB222131 LZPO01055098 OBS72520.1 JSUE03038719 JSUE03038720 JSUE03038721 AB083318
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000242457
+ More
UP000005203 UP000053105 UP000053825 UP000076502 UP000000311 UP000030742 UP000005205 UP000075809 UP000007266 UP000027135 UP000078540 UP000235965 UP000001554 UP000053097 UP000078492 UP000002279 UP000078541 UP000076858 UP000007755 UP000000305 UP000001811 UP000248480 UP000248481 UP000245341 UP000002358 UP000029965 UP000006813 UP000261680 UP000008225 UP000028761 UP000001595 UP000005447 UP000233140 UP000001519 UP000005640 UP000002277 UP000233080 UP000233040 UP000092124 UP000006718 UP000233100 UP000248482
UP000005203 UP000053105 UP000053825 UP000076502 UP000000311 UP000030742 UP000005205 UP000075809 UP000007266 UP000027135 UP000078540 UP000235965 UP000001554 UP000053097 UP000078492 UP000002279 UP000078541 UP000076858 UP000007755 UP000000305 UP000001811 UP000248480 UP000248481 UP000245341 UP000002358 UP000029965 UP000006813 UP000261680 UP000008225 UP000028761 UP000001595 UP000005447 UP000233140 UP000001519 UP000005640 UP000002277 UP000233080 UP000233040 UP000092124 UP000006718 UP000233100 UP000248482
Interpro
SUPFAM
SSF51182
SSF51182
Gene 3D
ProteinModelPortal
H9JAC8
A0A2A4JHC2
A0A2H1VVA5
A0A194R9L3
A0A194Q981
A0A212F4K2
+ More
A0A2A3E394 A0A088AHA0 A0A0M9A0D6 A0A0L7QST5 A0A154PCE3 A0A1B6FT62 A0A1B6MFM9 E2ARN2 A0A1B6EHC0 U4U4R5 A0A158NAH7 A0A151X534 D1ZZV1 A0A067QXA2 A0A0C9RX77 A0A151I5Q0 A0A2J7QHS1 A0A1Y1JXN4 C3ZF62 A0A026WK70 A0A195E4Y5 F6YAT8 A0A195EWE4 A0A164RYL5 A0A0P5I5N2 F4X239 A0A0P5QDA1 A0A0P6JWG5 A0A0P5PEQ5 A0A0P4XL22 E9GEW1 A0A2J7QHS8 A0A0P6BXW0 A0A0P5WY92 A0A0P5P0F0 A0A310SE35 G1SUK4 E9I9L0 A0A0P5P741 J3JX02 A0A2Y9DHN1 A0A2Y9HJA3 A0A2U3YUA8 A0A0P5ZL69 K7J6J4 V9KPF4 A0A0D9RPC6 G5AL67 U3CWX3 A0A384DIR6 A0A384DIV2 U3DUA7 A0A096NMS2 H2NNT1 H0VHT6 A0A2K5YTF3 G2HIF8 G3RFM0 P34949 H2Q9T4 A5A6K3 A0A2K5JTJ0 A0A2K5S6P1 A0A1A6H3X0 A0A1D5Q678 Q8HXX2 A0A2Y9K7X3
A0A2A3E394 A0A088AHA0 A0A0M9A0D6 A0A0L7QST5 A0A154PCE3 A0A1B6FT62 A0A1B6MFM9 E2ARN2 A0A1B6EHC0 U4U4R5 A0A158NAH7 A0A151X534 D1ZZV1 A0A067QXA2 A0A0C9RX77 A0A151I5Q0 A0A2J7QHS1 A0A1Y1JXN4 C3ZF62 A0A026WK70 A0A195E4Y5 F6YAT8 A0A195EWE4 A0A164RYL5 A0A0P5I5N2 F4X239 A0A0P5QDA1 A0A0P6JWG5 A0A0P5PEQ5 A0A0P4XL22 E9GEW1 A0A2J7QHS8 A0A0P6BXW0 A0A0P5WY92 A0A0P5P0F0 A0A310SE35 G1SUK4 E9I9L0 A0A0P5P741 J3JX02 A0A2Y9DHN1 A0A2Y9HJA3 A0A2U3YUA8 A0A0P5ZL69 K7J6J4 V9KPF4 A0A0D9RPC6 G5AL67 U3CWX3 A0A384DIR6 A0A384DIV2 U3DUA7 A0A096NMS2 H2NNT1 H0VHT6 A0A2K5YTF3 G2HIF8 G3RFM0 P34949 H2Q9T4 A5A6K3 A0A2K5JTJ0 A0A2K5S6P1 A0A1A6H3X0 A0A1D5Q678 Q8HXX2 A0A2Y9K7X3
PDB
5NW7
E-value=1.32085e-67,
Score=651
Ontologies
KEGG
PATHWAY
GO
PANTHER
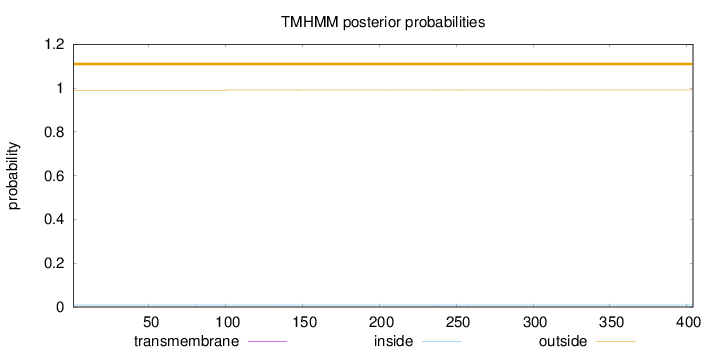
Topology
Subcellular location
Cytoplasm
Length:
404
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02466
Exp number, first 60 AAs:
0.00014
Total prob of N-in:
0.00954
outside
1 - 404
Population Genetic Test Statistics
Pi
205.839422
Theta
216.723345
Tajima's D
0.053594
CLR
0.044439
CSRT
0.378431078446078
Interpretation
Uncertain
