Gene
KWMTBOMO03339
Annotation
PREDICTED:_homeobox_protein_rough-like_[Papilio_polytes]
Full name
Homeobox protein rough
Transcription factor
Location in the cell
Nuclear Reliability : 3.666
Sequence
CDS
ATGGTGGCCCGGAGACGACGAAAAGAGAGTAGACCAAGACGACAGAGGACAACATTTTCAGCACATCAAACACTGCGATTAGAACTAGAGTACGCTAGAGGAGAATATGTTGCTAGAGCAAGAAGGTGTGAGTTAGCTTCGGCATTATCGTTAAGTGAAACCCAGGTTAAAATATGGTTTCAAAACAGAAGAGCCAAAGATAAAAGGATAGAGAAAGCGCAAATCGATCAACAATACAGGAATTTAGCTGCAGCCTCCGGTCTCTTCCCCACGGTACTCGGACCACCTTACTGCACCGCACCTGTTTGTCCATGTTTACCGGGGCCACAACCTTCAAGTATAGAGAAATAG
Protein
MVARRRRKESRPRRQRTTFSAHQTLRLELEYARGEYVARARRCELASALSLSETQVKIWFQNRRAKDKRIEKAQIDQQYRNLAAASGLFPTVLGPPYCTAPVCPCLPGPQPSSIEK
Summary
Description
Required to establish the unique cell identity of photoreceptors R2 and R5 and consequently for ommatidial assembly in the developing eye imaginal disk. Repression of expression in R8 photoreceptor by senseless (sens) is an essential mechanism of R8 cell fate determination.
Keywords
Complete proteome
Developmental protein
DNA-binding
Homeobox
Nucleus
Reference proteome
Sensory transduction
Transcription
Transcription regulation
Vision
Feature
chain Homeobox protein rough
Uniprot
A0A2A4IU88
A0A194RB78
A0A194Q350
A0A212F4M7
A0A336LQF5
A0A1B0GES9
+ More
A0A1A9W5V9 Q16E75 E9IEE7 A0A182XIB3 E2A2V9 B0W367 D6WB35 A0A158NMJ3 B4GYV9 A0A232FF53 B4PSB8 A0A0C9Q8C4 B4IC03 B4QWT1 K7JKN7 A0A182Y781 Q29BA4 A0A0L0CQH6 A7USV8 A0A182LFE3 A0A182Q4V6 N6TSE8 A0A182TTF8 P10181 B3P894 A0A1W4VL16 C7E1Y4 A0A0U2K775 K1Q149
A0A1A9W5V9 Q16E75 E9IEE7 A0A182XIB3 E2A2V9 B0W367 D6WB35 A0A158NMJ3 B4GYV9 A0A232FF53 B4PSB8 A0A0C9Q8C4 B4IC03 B4QWT1 K7JKN7 A0A182Y781 Q29BA4 A0A0L0CQH6 A7USV8 A0A182LFE3 A0A182Q4V6 N6TSE8 A0A182TTF8 P10181 B3P894 A0A1W4VL16 C7E1Y4 A0A0U2K775 K1Q149
Pubmed
EMBL
NWSH01007731
PCG62834.1
KQ460473
KPJ14525.1
KQ459551
KPI99982.1
+ More
AGBW02010347 OWR48673.1 UFQT01000111 SSX20180.1 CCAG010012581 CH478999 EAT32532.1 GL762591 EFZ21034.1 GL436217 EFN72233.1 DS231831 EDS31210.1 KQ971311 EEZ99430.2 ADTU01020387 ADTU01020388 CH479198 EDW27977.1 NNAY01000306 OXU29332.1 CM000160 EDW98580.1 GBYB01010478 JAG80245.1 CH480828 EDW45161.1 CM000364 EDX14598.1 CM000070 EAL27094.3 JRES01000062 KNC34501.1 AAAB01008859 EDO64243.2 AXCN02000061 APGK01053965 KB741239 KB632275 ENN72175.1 ERL91143.1 M23629 AE014297 BT022142 X12528 CH954182 EDV53498.1 GQ240850 ACT36592.1 KT984254 ALR88651.1 JH817886 EKC27628.1
AGBW02010347 OWR48673.1 UFQT01000111 SSX20180.1 CCAG010012581 CH478999 EAT32532.1 GL762591 EFZ21034.1 GL436217 EFN72233.1 DS231831 EDS31210.1 KQ971311 EEZ99430.2 ADTU01020387 ADTU01020388 CH479198 EDW27977.1 NNAY01000306 OXU29332.1 CM000160 EDW98580.1 GBYB01010478 JAG80245.1 CH480828 EDW45161.1 CM000364 EDX14598.1 CM000070 EAL27094.3 JRES01000062 KNC34501.1 AAAB01008859 EDO64243.2 AXCN02000061 APGK01053965 KB741239 KB632275 ENN72175.1 ERL91143.1 M23629 AE014297 BT022142 X12528 CH954182 EDV53498.1 GQ240850 ACT36592.1 KT984254 ALR88651.1 JH817886 EKC27628.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000092444
UP000091820
+ More
UP000008820 UP000076407 UP000000311 UP000002320 UP000007266 UP000005205 UP000008744 UP000215335 UP000002282 UP000001292 UP000000304 UP000002358 UP000076408 UP000001819 UP000037069 UP000007062 UP000075882 UP000075886 UP000019118 UP000030742 UP000075902 UP000000803 UP000008711 UP000192221 UP000005408
UP000008820 UP000076407 UP000000311 UP000002320 UP000007266 UP000005205 UP000008744 UP000215335 UP000002282 UP000001292 UP000000304 UP000002358 UP000076408 UP000001819 UP000037069 UP000007062 UP000075882 UP000075886 UP000019118 UP000030742 UP000075902 UP000000803 UP000008711 UP000192221 UP000005408
PRIDE
Pfam
PF00046 Homeodomain
Interpro
SUPFAM
SSF46689
SSF46689
ProteinModelPortal
A0A2A4IU88
A0A194RB78
A0A194Q350
A0A212F4M7
A0A336LQF5
A0A1B0GES9
+ More
A0A1A9W5V9 Q16E75 E9IEE7 A0A182XIB3 E2A2V9 B0W367 D6WB35 A0A158NMJ3 B4GYV9 A0A232FF53 B4PSB8 A0A0C9Q8C4 B4IC03 B4QWT1 K7JKN7 A0A182Y781 Q29BA4 A0A0L0CQH6 A7USV8 A0A182LFE3 A0A182Q4V6 N6TSE8 A0A182TTF8 P10181 B3P894 A0A1W4VL16 C7E1Y4 A0A0U2K775 K1Q149
A0A1A9W5V9 Q16E75 E9IEE7 A0A182XIB3 E2A2V9 B0W367 D6WB35 A0A158NMJ3 B4GYV9 A0A232FF53 B4PSB8 A0A0C9Q8C4 B4IC03 B4QWT1 K7JKN7 A0A182Y781 Q29BA4 A0A0L0CQH6 A7USV8 A0A182LFE3 A0A182Q4V6 N6TSE8 A0A182TTF8 P10181 B3P894 A0A1W4VL16 C7E1Y4 A0A0U2K775 K1Q149
PDB
6ES2
E-value=1.56904e-10,
Score=152
Ontologies
GO
Topology
Subcellular location
Nucleus
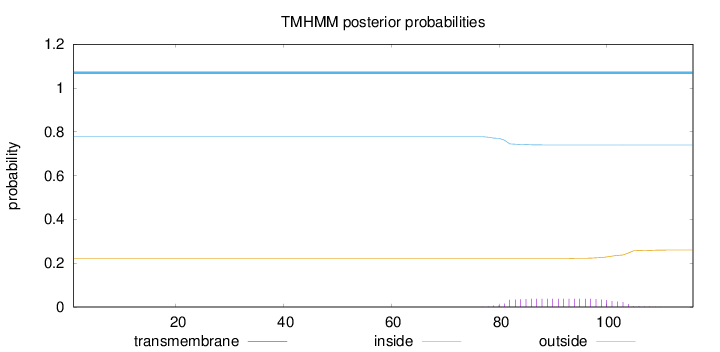
Length:
116
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.81899
Exp number, first 60 AAs:
0.00034
Total prob of N-in:
0.77775
inside
1 - 116
Population Genetic Test Statistics
Pi
182.902299
Theta
123.158306
Tajima's D
1.523193
CLR
0
CSRT
0.794010299485026
Interpretation
Uncertain
