Gene
KWMTBOMO03335
Pre Gene Modal
BGIBMGA006406
Annotation
seminal_fluid_protein_CSSFP002_[Chilo_suppressalis]
Location in the cell
Extracellular Reliability : 1.423 Nuclear Reliability : 1.412
Sequence
CDS
ATGGAGTGCGGTGGCGCCAACCAAGAGAACCGTATCGTGGGTGGGATGCCAGCCAGCTCAAACCGGTACCCGTGGATGGCCAGAATCGTTTACGACGGACAGTTCCACTGTGGAGCCTCAGTTCTCACCAAGGAATACGTGTTAACAGCAGCGCATTGTGTCAAGAAATTAAAAAGATCAAAGATCCGGGTGATCCTCGGAGATCACGACCAGACGATCACGTCCGAGAGTGCGGCCATCATGCGGGCGGTCTCTGCGATTGTCCGCCACCGTAGCTTCGACACTGAATCATACAATAACGATATAGCACTACTAAAGTTGAGAAAACCCGTGAATTTCTCTAAAATCATCAAGCCCGTTTGTTTGCCCCCTCCAAGTATTGACCCAGCTGGAAAAGAAGGTGTAGTGGTGGGATGGGGGCGAACATCCGAGGGGGGTCAATTACCCGCAGTTGTACAGGAGGTTCGCGTGCCCATTCTATCACTGAATCAGTGTCGAGGATTGAAATACCGTGCATCTAAGATCACTAACAACATGCTGTGTGCGGGCAGGGCGTCAACCGATTCTTGTCAAGGAGACAGCGGTGGGCCTTTGCTTCTTCAGCAAGGGGACAAGTATCAAATTGTTGGCATAGTGTCATGGGGAGTCGGTTGCGGTCGACCGGGATACCCGGGAGTCTACACGAGGATCACTCGATACCTACCCTGGCTTAAAGCAAACCTTCGAGACACATTAAATTAA
Protein
MECGGANQENRIVGGMPASSNRYPWMARIVYDGQFHCGASVLTKEYVLTAAHCVKKLKRSKIRVILGDHDQTITSESAAIMRAVSAIVRHRSFDTESYNNDIALLKLRKPVNFSKIIKPVCLPPPSIDPAGKEGVVVGWGRTSEGGQLPAVVQEVRVPILSLNQCRGLKYRASKITNNMLCAGRASTDSCQGDSGGPLLLQQGDKYQIVGIVSWGVGCGRPGYPGVYTRITRYLPWLKANLRDTLN
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
A0A2A4IT33
G9F9F2
A0A194R9L8
A0A212F4L7
A0A2H1VW37
F7IUA2
+ More
A0A1S4GM14 A0A182X2K6 A0A182M392 A0A1S4FGP8 Q171W1 Q17039 A0A182PQT1 A0A182K8A2 W5JJI7 A0A182T0F3 A0A182J3Z9 A0A084WT72 B0WAI9 D6W9N9 A0A139WME7 A0A0C9QMS3 A0A0C9RD19 A0A0A1WLN7 F4X2V2 B4PUE3 A0A195BW73 A0A182RDY5 A0A0C9R9I2 A0A195EK23 K7J296 A0A0K8VUH8 W8CC89 A0A151WZG8 I5AP59 A0A1W4VKZ7 J3JTS4 B4K4E2 E1JIW3 B3M1Y7 A0A3B0K5F5 A0A0N0BDH6 E0VW11 A0A068F4F5 A0A2A3E7J8 N6UAG3 A0A1I8PFM9 A0A1I8PFE2 A0A1B0FRC3 A0A1V1FVE0 A0A087ZRJ0 A0A1A9ZZP8 A0A1A9UZS8 A0A1B0C2U8 A0A1A9XN15 A0A1A9W6L8 A0A026WAC6 A0A067QVB6 A0A182N9T5 A0A3L8E3C9 A0A182Y8L3 A0A3Q0IU66 A0A195F5W7 E2AFY8 A0A0L7QZB2 A0A182QIC4 B4MC85 B4NJM4 B4GE38 B4QUA8 B4IJF3 Q9VBY4 B3P6S7 A0A1I8PFL2 B4JHT1 J9JS49 A0A2S2NYR9 A0A1J1IYB5 A0A2H8TI15 A0A194Q371 A0A182TX81 A0A2S2QSA4 A0A182F8Y9 A0A182W3L9 A0A2J7Q479 E2BDT6 A0A2R7WXS3 A0A182HYU9 A0A182KZD9 A0A1B0D036 A0A195D4U7 A0A2A3E772 A0A195F5K4 A0A195EJ73 A0A087ZRI1 A0A158P398 A0A026WB88 E0VW09
A0A1S4GM14 A0A182X2K6 A0A182M392 A0A1S4FGP8 Q171W1 Q17039 A0A182PQT1 A0A182K8A2 W5JJI7 A0A182T0F3 A0A182J3Z9 A0A084WT72 B0WAI9 D6W9N9 A0A139WME7 A0A0C9QMS3 A0A0C9RD19 A0A0A1WLN7 F4X2V2 B4PUE3 A0A195BW73 A0A182RDY5 A0A0C9R9I2 A0A195EK23 K7J296 A0A0K8VUH8 W8CC89 A0A151WZG8 I5AP59 A0A1W4VKZ7 J3JTS4 B4K4E2 E1JIW3 B3M1Y7 A0A3B0K5F5 A0A0N0BDH6 E0VW11 A0A068F4F5 A0A2A3E7J8 N6UAG3 A0A1I8PFM9 A0A1I8PFE2 A0A1B0FRC3 A0A1V1FVE0 A0A087ZRJ0 A0A1A9ZZP8 A0A1A9UZS8 A0A1B0C2U8 A0A1A9XN15 A0A1A9W6L8 A0A026WAC6 A0A067QVB6 A0A182N9T5 A0A3L8E3C9 A0A182Y8L3 A0A3Q0IU66 A0A195F5W7 E2AFY8 A0A0L7QZB2 A0A182QIC4 B4MC85 B4NJM4 B4GE38 B4QUA8 B4IJF3 Q9VBY4 B3P6S7 A0A1I8PFL2 B4JHT1 J9JS49 A0A2S2NYR9 A0A1J1IYB5 A0A2H8TI15 A0A194Q371 A0A182TX81 A0A2S2QSA4 A0A182F8Y9 A0A182W3L9 A0A2J7Q479 E2BDT6 A0A2R7WXS3 A0A182HYU9 A0A182KZD9 A0A1B0D036 A0A195D4U7 A0A2A3E772 A0A195F5K4 A0A195EJ73 A0A087ZRI1 A0A158P398 A0A026WB88 E0VW09
Pubmed
26354079
22118469
12364791
17510324
8630536
20920257
+ More
23761445 24438588 18362917 19820115 25830018 21719571 17994087 17550304 20075255 24495485 15632085 22516182 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20566863 24952583 28410430 24508170 24845553 30249741 25244985 20798317 18057021 26109357 26109356 20966253 21347285
23761445 24438588 18362917 19820115 25830018 21719571 17994087 17550304 20075255 24495485 15632085 22516182 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20566863 24952583 28410430 24508170 24845553 30249741 25244985 20798317 18057021 26109357 26109356 20966253 21347285
EMBL
NWSH01007731
PCG62829.1
JN033697
AEW46850.1
KQ460473
KPJ14528.1
+ More
AGBW02010347 OWR48670.1 ODYU01004780 SOQ45018.1 AAAB01008898 EAA09162.5 AXCM01002956 CH477445 EAT40775.1 Z49813 CAA89967.1 ADMH02001350 ETN62924.1 ATLV01026864 KE525420 KFB53416.1 DS231872 EDS41412.1 KQ971312 EEZ99214.2 KYB29219.1 GBYB01004789 JAG74556.1 GBYB01004791 JAG74558.1 GBXI01014535 JAC99756.1 GL888593 EGI59207.1 CM000160 EDW98830.2 KQ976403 KYM92218.1 GBYB01004790 JAG74557.1 KQ978801 KYN28214.1 AAZX01018692 GDHF01010114 JAI42200.1 GAMC01004116 JAC02440.1 KQ982649 KYQ53176.1 CM000070 EIM52744.2 BT126633 KB632307 AEE61596.1 ERL91907.1 CH933806 EDW13894.2 AE014297 ACZ95012.2 CH902617 EDV42247.2 OUUW01000007 SPP83250.1 KQ435859 KOX70461.1 DS235816 EEB17567.1 KJ512103 AID60326.1 KZ288360 PBC27021.1 APGK01042919 APGK01042920 KB741010 ENN75592.1 CCAG010008016 FX985325 BAX07338.1 JXJN01024710 KK107304 EZA52933.1 KK853216 KDR09771.1 QOIP01000001 RLU27156.1 KQ981805 KYN35464.1 GL439184 EFN67638.1 KQ414681 KOC63969.1 AXCN02001200 CH940657 EDW63167.1 KRF80937.1 KRF80938.1 CH964272 EDW83948.1 CH479182 EDW33873.1 CM000364 EDX14345.1 CH480848 EDW51144.1 AY061333 AAF56393.1 AAL28881.1 CH954182 EDV53747.1 CH916369 EDV93920.1 ABLF02030881 GGMR01009655 MBY22274.1 CVRI01000064 CRL05275.1 GFXV01001962 MBW13767.1 KQ459551 KPI99986.1 GGMS01010809 MBY80012.1 NEVH01018390 PNF23380.1 GL447689 EFN86142.1 KK855905 PTY24326.1 APCN01005409 AJVK01009807 KQ976842 KYN07908.1 PBC27019.1 KYN35462.1 KYN28211.1 ADTU01007949 EZA52931.1 RLU26344.1 EEB17565.1
AGBW02010347 OWR48670.1 ODYU01004780 SOQ45018.1 AAAB01008898 EAA09162.5 AXCM01002956 CH477445 EAT40775.1 Z49813 CAA89967.1 ADMH02001350 ETN62924.1 ATLV01026864 KE525420 KFB53416.1 DS231872 EDS41412.1 KQ971312 EEZ99214.2 KYB29219.1 GBYB01004789 JAG74556.1 GBYB01004791 JAG74558.1 GBXI01014535 JAC99756.1 GL888593 EGI59207.1 CM000160 EDW98830.2 KQ976403 KYM92218.1 GBYB01004790 JAG74557.1 KQ978801 KYN28214.1 AAZX01018692 GDHF01010114 JAI42200.1 GAMC01004116 JAC02440.1 KQ982649 KYQ53176.1 CM000070 EIM52744.2 BT126633 KB632307 AEE61596.1 ERL91907.1 CH933806 EDW13894.2 AE014297 ACZ95012.2 CH902617 EDV42247.2 OUUW01000007 SPP83250.1 KQ435859 KOX70461.1 DS235816 EEB17567.1 KJ512103 AID60326.1 KZ288360 PBC27021.1 APGK01042919 APGK01042920 KB741010 ENN75592.1 CCAG010008016 FX985325 BAX07338.1 JXJN01024710 KK107304 EZA52933.1 KK853216 KDR09771.1 QOIP01000001 RLU27156.1 KQ981805 KYN35464.1 GL439184 EFN67638.1 KQ414681 KOC63969.1 AXCN02001200 CH940657 EDW63167.1 KRF80937.1 KRF80938.1 CH964272 EDW83948.1 CH479182 EDW33873.1 CM000364 EDX14345.1 CH480848 EDW51144.1 AY061333 AAF56393.1 AAL28881.1 CH954182 EDV53747.1 CH916369 EDV93920.1 ABLF02030881 GGMR01009655 MBY22274.1 CVRI01000064 CRL05275.1 GFXV01001962 MBW13767.1 KQ459551 KPI99986.1 GGMS01010809 MBY80012.1 NEVH01018390 PNF23380.1 GL447689 EFN86142.1 KK855905 PTY24326.1 APCN01005409 AJVK01009807 KQ976842 KYN07908.1 PBC27019.1 KYN35462.1 KYN28211.1 ADTU01007949 EZA52931.1 RLU26344.1 EEB17565.1
Proteomes
UP000218220
UP000053240
UP000007151
UP000007062
UP000076407
UP000075883
+ More
UP000008820 UP000075885 UP000075881 UP000000673 UP000075901 UP000075880 UP000030765 UP000002320 UP000007266 UP000007755 UP000002282 UP000078540 UP000075900 UP000078492 UP000002358 UP000075809 UP000001819 UP000192221 UP000030742 UP000009192 UP000000803 UP000007801 UP000268350 UP000053105 UP000009046 UP000242457 UP000019118 UP000095300 UP000092444 UP000005203 UP000092445 UP000078200 UP000092460 UP000092443 UP000091820 UP000053097 UP000027135 UP000075884 UP000279307 UP000076408 UP000079169 UP000078541 UP000000311 UP000053825 UP000075886 UP000008792 UP000007798 UP000008744 UP000000304 UP000001292 UP000008711 UP000001070 UP000007819 UP000183832 UP000053268 UP000075902 UP000069272 UP000075920 UP000235965 UP000008237 UP000075840 UP000075882 UP000092462 UP000078542 UP000005205
UP000008820 UP000075885 UP000075881 UP000000673 UP000075901 UP000075880 UP000030765 UP000002320 UP000007266 UP000007755 UP000002282 UP000078540 UP000075900 UP000078492 UP000002358 UP000075809 UP000001819 UP000192221 UP000030742 UP000009192 UP000000803 UP000007801 UP000268350 UP000053105 UP000009046 UP000242457 UP000019118 UP000095300 UP000092444 UP000005203 UP000092445 UP000078200 UP000092460 UP000092443 UP000091820 UP000053097 UP000027135 UP000075884 UP000279307 UP000076408 UP000079169 UP000078541 UP000000311 UP000053825 UP000075886 UP000008792 UP000007798 UP000008744 UP000000304 UP000001292 UP000008711 UP000001070 UP000007819 UP000183832 UP000053268 UP000075902 UP000069272 UP000075920 UP000235965 UP000008237 UP000075840 UP000075882 UP000092462 UP000078542 UP000005205
PRIDE
Pfam
PF00089 Trypsin
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
A0A2A4IT33
G9F9F2
A0A194R9L8
A0A212F4L7
A0A2H1VW37
F7IUA2
+ More
A0A1S4GM14 A0A182X2K6 A0A182M392 A0A1S4FGP8 Q171W1 Q17039 A0A182PQT1 A0A182K8A2 W5JJI7 A0A182T0F3 A0A182J3Z9 A0A084WT72 B0WAI9 D6W9N9 A0A139WME7 A0A0C9QMS3 A0A0C9RD19 A0A0A1WLN7 F4X2V2 B4PUE3 A0A195BW73 A0A182RDY5 A0A0C9R9I2 A0A195EK23 K7J296 A0A0K8VUH8 W8CC89 A0A151WZG8 I5AP59 A0A1W4VKZ7 J3JTS4 B4K4E2 E1JIW3 B3M1Y7 A0A3B0K5F5 A0A0N0BDH6 E0VW11 A0A068F4F5 A0A2A3E7J8 N6UAG3 A0A1I8PFM9 A0A1I8PFE2 A0A1B0FRC3 A0A1V1FVE0 A0A087ZRJ0 A0A1A9ZZP8 A0A1A9UZS8 A0A1B0C2U8 A0A1A9XN15 A0A1A9W6L8 A0A026WAC6 A0A067QVB6 A0A182N9T5 A0A3L8E3C9 A0A182Y8L3 A0A3Q0IU66 A0A195F5W7 E2AFY8 A0A0L7QZB2 A0A182QIC4 B4MC85 B4NJM4 B4GE38 B4QUA8 B4IJF3 Q9VBY4 B3P6S7 A0A1I8PFL2 B4JHT1 J9JS49 A0A2S2NYR9 A0A1J1IYB5 A0A2H8TI15 A0A194Q371 A0A182TX81 A0A2S2QSA4 A0A182F8Y9 A0A182W3L9 A0A2J7Q479 E2BDT6 A0A2R7WXS3 A0A182HYU9 A0A182KZD9 A0A1B0D036 A0A195D4U7 A0A2A3E772 A0A195F5K4 A0A195EJ73 A0A087ZRI1 A0A158P398 A0A026WB88 E0VW09
A0A1S4GM14 A0A182X2K6 A0A182M392 A0A1S4FGP8 Q171W1 Q17039 A0A182PQT1 A0A182K8A2 W5JJI7 A0A182T0F3 A0A182J3Z9 A0A084WT72 B0WAI9 D6W9N9 A0A139WME7 A0A0C9QMS3 A0A0C9RD19 A0A0A1WLN7 F4X2V2 B4PUE3 A0A195BW73 A0A182RDY5 A0A0C9R9I2 A0A195EK23 K7J296 A0A0K8VUH8 W8CC89 A0A151WZG8 I5AP59 A0A1W4VKZ7 J3JTS4 B4K4E2 E1JIW3 B3M1Y7 A0A3B0K5F5 A0A0N0BDH6 E0VW11 A0A068F4F5 A0A2A3E7J8 N6UAG3 A0A1I8PFM9 A0A1I8PFE2 A0A1B0FRC3 A0A1V1FVE0 A0A087ZRJ0 A0A1A9ZZP8 A0A1A9UZS8 A0A1B0C2U8 A0A1A9XN15 A0A1A9W6L8 A0A026WAC6 A0A067QVB6 A0A182N9T5 A0A3L8E3C9 A0A182Y8L3 A0A3Q0IU66 A0A195F5W7 E2AFY8 A0A0L7QZB2 A0A182QIC4 B4MC85 B4NJM4 B4GE38 B4QUA8 B4IJF3 Q9VBY4 B3P6S7 A0A1I8PFL2 B4JHT1 J9JS49 A0A2S2NYR9 A0A1J1IYB5 A0A2H8TI15 A0A194Q371 A0A182TX81 A0A2S2QSA4 A0A182F8Y9 A0A182W3L9 A0A2J7Q479 E2BDT6 A0A2R7WXS3 A0A182HYU9 A0A182KZD9 A0A1B0D036 A0A195D4U7 A0A2A3E772 A0A195F5K4 A0A195EJ73 A0A087ZRI1 A0A158P398 A0A026WB88 E0VW09
PDB
3W94
E-value=3.15662e-46,
Score=464
Ontologies
GO
Topology
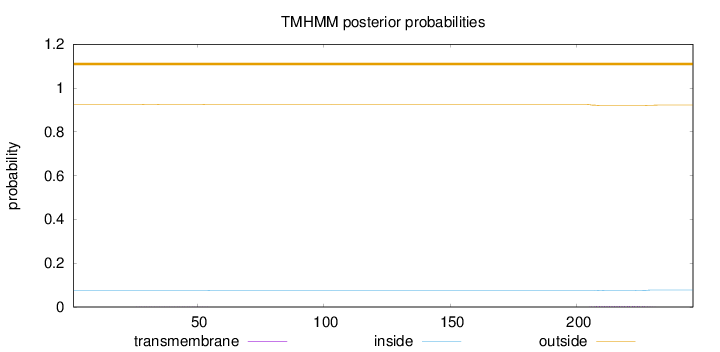
Length:
246
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10537
Exp number, first 60 AAs:
0.01281
Total prob of N-in:
0.07562
outside
1 - 246
Population Genetic Test Statistics
Pi
170.987369
Theta
155.882285
Tajima's D
-1.382566
CLR
0.621304
CSRT
0.0775461226938653
Interpretation
Uncertain
