Gene
KWMTBOMO03330
Pre Gene Modal
BGIBMGA006471
Annotation
stearoyl-CoA_desaturase_5-like_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.861
Sequence
CDS
ATGTATGCAGGGAGCACAGGCGTTACAGTCGGTGCTCATCGACTCTTCACTCATAAATCATTCAAAGCAACACCGCTTCTCAAAGTTTTTCTCCTTTGCCTACAAACCATAGCAGGACAGAATTCGACGTTCATTTGGGTTCGCGATCATCGTCTCCACCACCGCTACTCCGATACCGATGCCGACCCACATAATTCTAAGAGAGGCTTCTTCTTTTGCCACATTGGATGGCTAATGATGAAAAAACATCCATATGTTATTGAATTGGGAAGAAGAATCGATATGAGTGACCTTCAAGCGGATAAAATGATAATGTTCCAGAAAAAGTAA
Protein
MYAGSTGVTVGAHRLFTHKSFKATPLLKVFLLCLQTIAGQNSTFIWVRDHRLHHRYSDTDADPHNSKRGFFFCHIGWLMMKKHPYVIELGRRIDMSDLQADKMIMFQKK
Summary
Cofactor
Fe cation
Fe(2+)
Fe(2+)
Similarity
Belongs to the fatty acid desaturase type 1 family.
Uniprot
T1RTH0
T1RTE2
H9JAC6
T1RTG1
A0A2H1WSU7
A0A194Q989
+ More
A0A194RF26 A0A2H1V292 A0A2A4IXV6 A0A1L8D6C1 A0A212F4K6 A0A182JUS5 T1IDC7 A0A182PQT3 A0A0L7KSL0 A0A182W3M1 A0A182RDY4 A0A182U951 A0A182F8Y7 A0A182N9T4 A0A1B6FRL5 E0VW13 A0A182VM37 A0A2C9H8A7 A0A1B6KF34 A0A2C9GQA8 A0A182KZD7 Q7PQ92 A0A182Y8L2 W5JFU6 A0A1B0D038 A0A1B0CZP5 A0A182IMC8 A0A182Q8Q9 A0A084WT65 A0A182MAQ2 T1I088 A0A0T6AYP7 A0A182SRM0 A0A146L0C4 A0A2R7VQY9 A0A1J1IZY5 D6W9N6 A0A096XDB4 Q171V8 A0A336LJV2 A4KWW3 A4ZKB7 H9JAC5 Q8WSZ4 Q8WQP9 A4ZKB8 A0A1B0CZP4 Q14TE5 B0WAJ2 A0A1B0CZP1 T1IDE9 A0A1Y1LII3 A0A1W4WVL1 A0A182PS55 A0A2S2QMZ6 W5JSC3 A0A023EUP4 A0A3M6TZ36 N6UAX5 A0A023ERW8 A0A0F8B2S9 A0A1J1J657 T1PQM9 E2BGQ2 A0A336MAE4 A0A3B4Y3V9 A0A3B4U151 A0A1S4EFC2 Q173B1 A0A0K8S5D1 A0A182IPM3 A0A182TJZ6 A0A023ERW2 A0A1S3D6B4 A0A1S3D7T3 A0A0P6J0V5 A0A0A9XP67 A0A1S4G6X1 A0A2S2QTL0 A0A1S4ECH4 M1FXD6 A0A182R814 T1I119 U5EY18 A0A182T7Z9 A0A2D4KYW2 A0A2S2QIW8 A0A1B0DIJ1 A0A182KWY6 A0A182HJS3 Q7QDJ5 A0A182XK65 A0A182V4X1
A0A194RF26 A0A2H1V292 A0A2A4IXV6 A0A1L8D6C1 A0A212F4K6 A0A182JUS5 T1IDC7 A0A182PQT3 A0A0L7KSL0 A0A182W3M1 A0A182RDY4 A0A182U951 A0A182F8Y7 A0A182N9T4 A0A1B6FRL5 E0VW13 A0A182VM37 A0A2C9H8A7 A0A1B6KF34 A0A2C9GQA8 A0A182KZD7 Q7PQ92 A0A182Y8L2 W5JFU6 A0A1B0D038 A0A1B0CZP5 A0A182IMC8 A0A182Q8Q9 A0A084WT65 A0A182MAQ2 T1I088 A0A0T6AYP7 A0A182SRM0 A0A146L0C4 A0A2R7VQY9 A0A1J1IZY5 D6W9N6 A0A096XDB4 Q171V8 A0A336LJV2 A4KWW3 A4ZKB7 H9JAC5 Q8WSZ4 Q8WQP9 A4ZKB8 A0A1B0CZP4 Q14TE5 B0WAJ2 A0A1B0CZP1 T1IDE9 A0A1Y1LII3 A0A1W4WVL1 A0A182PS55 A0A2S2QMZ6 W5JSC3 A0A023EUP4 A0A3M6TZ36 N6UAX5 A0A023ERW8 A0A0F8B2S9 A0A1J1J657 T1PQM9 E2BGQ2 A0A336MAE4 A0A3B4Y3V9 A0A3B4U151 A0A1S4EFC2 Q173B1 A0A0K8S5D1 A0A182IPM3 A0A182TJZ6 A0A023ERW2 A0A1S3D6B4 A0A1S3D7T3 A0A0P6J0V5 A0A0A9XP67 A0A1S4G6X1 A0A2S2QTL0 A0A1S4ECH4 M1FXD6 A0A182R814 T1I119 U5EY18 A0A182T7Z9 A0A2D4KYW2 A0A2S2QIW8 A0A1B0DIJ1 A0A182KWY6 A0A182HJS3 Q7QDJ5 A0A182XK65 A0A182V4X1
Pubmed
EMBL
JX531671
AGO45856.1
JX531672
AGO45857.1
BABH01009091
JX531673
+ More
AGO45858.1 ODYU01010795 SOQ56118.1 KQ459551 KPI99990.1 KQ460473 KPJ14531.1 ODYU01000347 SOQ34951.1 NWSH01004663 PCG64797.1 GEYN01000127 JAV02002.1 AGBW02010347 OWR48667.1 ACPB03002937 JTDY01006147 KOB66242.1 GECZ01016924 JAS52845.1 DS235816 EEB17569.1 GEBQ01029895 JAT10082.1 APCN01005409 AAAB01008898 EAA09155.6 EGK97208.1 ADMH02001350 ETN62926.1 AJVK01009807 AJVK01020909 AXCN02001200 ATLV01026860 KE525420 KFB53409.1 AXCM01000318 ACPB03011802 LJIG01022516 KRT80162.1 GDHC01018259 JAQ00370.1 KK854022 PTY09558.1 CVRI01000064 CRL05274.1 KQ971312 EEZ98125.1 KF576671 AHH30812.1 CH477445 EAT40778.1 UFQS01006037 UFQT01006037 SSX16909.1 SSX36077.1 EF125923 EF125924 ABO43931.1 ABO43932.1 EF125925 ABO43933.1 BABH01009086 BABH01009087 BABH01009088 BABH01009089 AF441220 EU350085 EU350086 AAL35330.1 ACA81689.1 ACA81690.1 AY062023 AAL35746.2 EF125926 ABO43934.1 AJVK01020908 AB264085 BAE97679.1 DS231872 EDS41415.1 AJVK01020907 GEZM01060888 GEZM01060887 JAV70837.1 GGMS01009319 MBY78522.1 ADMH02000313 ETN67016.1 GAPW01000906 JAC12692.1 RCHS01002593 RMX46715.1 APGK01042247 APGK01042248 APGK01042249 APGK01042250 APGK01042251 KB741002 KB632309 ENN75787.1 ERL92046.1 GAPW01002159 JAC11439.1 KP731998 KQ041834 ALL96430.1 KKF21888.1 CVRI01000074 CRL07879.1 KA650423 AFP65052.1 GL448204 EFN85122.1 UFQT01000797 SSX27312.1 CH477428 EAT41117.1 GBRD01017443 GBRD01017442 JAG48384.1 GAPW01001631 JAC11967.1 GDUN01000309 JAN95610.1 GBHO01040158 GBHO01040157 GBHO01040154 GBHO01021850 JAG03446.1 JAG03447.1 JAG03450.1 JAG21754.1 GGMS01011770 MBY80973.1 JQ693685 AFV15454.1 ACPB03020384 GANO01002241 JAB57630.1 IACL01099204 LAB13896.1 GGMS01008481 MBY77684.1 AJVK01062643 APCN01002208 AAAB01008851 EAA07365.5
AGO45858.1 ODYU01010795 SOQ56118.1 KQ459551 KPI99990.1 KQ460473 KPJ14531.1 ODYU01000347 SOQ34951.1 NWSH01004663 PCG64797.1 GEYN01000127 JAV02002.1 AGBW02010347 OWR48667.1 ACPB03002937 JTDY01006147 KOB66242.1 GECZ01016924 JAS52845.1 DS235816 EEB17569.1 GEBQ01029895 JAT10082.1 APCN01005409 AAAB01008898 EAA09155.6 EGK97208.1 ADMH02001350 ETN62926.1 AJVK01009807 AJVK01020909 AXCN02001200 ATLV01026860 KE525420 KFB53409.1 AXCM01000318 ACPB03011802 LJIG01022516 KRT80162.1 GDHC01018259 JAQ00370.1 KK854022 PTY09558.1 CVRI01000064 CRL05274.1 KQ971312 EEZ98125.1 KF576671 AHH30812.1 CH477445 EAT40778.1 UFQS01006037 UFQT01006037 SSX16909.1 SSX36077.1 EF125923 EF125924 ABO43931.1 ABO43932.1 EF125925 ABO43933.1 BABH01009086 BABH01009087 BABH01009088 BABH01009089 AF441220 EU350085 EU350086 AAL35330.1 ACA81689.1 ACA81690.1 AY062023 AAL35746.2 EF125926 ABO43934.1 AJVK01020908 AB264085 BAE97679.1 DS231872 EDS41415.1 AJVK01020907 GEZM01060888 GEZM01060887 JAV70837.1 GGMS01009319 MBY78522.1 ADMH02000313 ETN67016.1 GAPW01000906 JAC12692.1 RCHS01002593 RMX46715.1 APGK01042247 APGK01042248 APGK01042249 APGK01042250 APGK01042251 KB741002 KB632309 ENN75787.1 ERL92046.1 GAPW01002159 JAC11439.1 KP731998 KQ041834 ALL96430.1 KKF21888.1 CVRI01000074 CRL07879.1 KA650423 AFP65052.1 GL448204 EFN85122.1 UFQT01000797 SSX27312.1 CH477428 EAT41117.1 GBRD01017443 GBRD01017442 JAG48384.1 GAPW01001631 JAC11967.1 GDUN01000309 JAN95610.1 GBHO01040158 GBHO01040157 GBHO01040154 GBHO01021850 JAG03446.1 JAG03447.1 JAG03450.1 JAG21754.1 GGMS01011770 MBY80973.1 JQ693685 AFV15454.1 ACPB03020384 GANO01002241 JAB57630.1 IACL01099204 LAB13896.1 GGMS01008481 MBY77684.1 AJVK01062643 APCN01002208 AAAB01008851 EAA07365.5
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007151
UP000075881
+ More
UP000015103 UP000075885 UP000037510 UP000075920 UP000075900 UP000075902 UP000069272 UP000075884 UP000009046 UP000075903 UP000076407 UP000075840 UP000075882 UP000007062 UP000076408 UP000000673 UP000092462 UP000075880 UP000075886 UP000030765 UP000075883 UP000075901 UP000183832 UP000007266 UP000008820 UP000002320 UP000192223 UP000275408 UP000019118 UP000030742 UP000008237 UP000261360 UP000261420 UP000079169
UP000015103 UP000075885 UP000037510 UP000075920 UP000075900 UP000075902 UP000069272 UP000075884 UP000009046 UP000075903 UP000076407 UP000075840 UP000075882 UP000007062 UP000076408 UP000000673 UP000092462 UP000075880 UP000075886 UP000030765 UP000075883 UP000075901 UP000183832 UP000007266 UP000008820 UP000002320 UP000192223 UP000275408 UP000019118 UP000030742 UP000008237 UP000261360 UP000261420 UP000079169
Pfam
PF00487 FA_desaturase
Interpro
ProteinModelPortal
T1RTH0
T1RTE2
H9JAC6
T1RTG1
A0A2H1WSU7
A0A194Q989
+ More
A0A194RF26 A0A2H1V292 A0A2A4IXV6 A0A1L8D6C1 A0A212F4K6 A0A182JUS5 T1IDC7 A0A182PQT3 A0A0L7KSL0 A0A182W3M1 A0A182RDY4 A0A182U951 A0A182F8Y7 A0A182N9T4 A0A1B6FRL5 E0VW13 A0A182VM37 A0A2C9H8A7 A0A1B6KF34 A0A2C9GQA8 A0A182KZD7 Q7PQ92 A0A182Y8L2 W5JFU6 A0A1B0D038 A0A1B0CZP5 A0A182IMC8 A0A182Q8Q9 A0A084WT65 A0A182MAQ2 T1I088 A0A0T6AYP7 A0A182SRM0 A0A146L0C4 A0A2R7VQY9 A0A1J1IZY5 D6W9N6 A0A096XDB4 Q171V8 A0A336LJV2 A4KWW3 A4ZKB7 H9JAC5 Q8WSZ4 Q8WQP9 A4ZKB8 A0A1B0CZP4 Q14TE5 B0WAJ2 A0A1B0CZP1 T1IDE9 A0A1Y1LII3 A0A1W4WVL1 A0A182PS55 A0A2S2QMZ6 W5JSC3 A0A023EUP4 A0A3M6TZ36 N6UAX5 A0A023ERW8 A0A0F8B2S9 A0A1J1J657 T1PQM9 E2BGQ2 A0A336MAE4 A0A3B4Y3V9 A0A3B4U151 A0A1S4EFC2 Q173B1 A0A0K8S5D1 A0A182IPM3 A0A182TJZ6 A0A023ERW2 A0A1S3D6B4 A0A1S3D7T3 A0A0P6J0V5 A0A0A9XP67 A0A1S4G6X1 A0A2S2QTL0 A0A1S4ECH4 M1FXD6 A0A182R814 T1I119 U5EY18 A0A182T7Z9 A0A2D4KYW2 A0A2S2QIW8 A0A1B0DIJ1 A0A182KWY6 A0A182HJS3 Q7QDJ5 A0A182XK65 A0A182V4X1
A0A194RF26 A0A2H1V292 A0A2A4IXV6 A0A1L8D6C1 A0A212F4K6 A0A182JUS5 T1IDC7 A0A182PQT3 A0A0L7KSL0 A0A182W3M1 A0A182RDY4 A0A182U951 A0A182F8Y7 A0A182N9T4 A0A1B6FRL5 E0VW13 A0A182VM37 A0A2C9H8A7 A0A1B6KF34 A0A2C9GQA8 A0A182KZD7 Q7PQ92 A0A182Y8L2 W5JFU6 A0A1B0D038 A0A1B0CZP5 A0A182IMC8 A0A182Q8Q9 A0A084WT65 A0A182MAQ2 T1I088 A0A0T6AYP7 A0A182SRM0 A0A146L0C4 A0A2R7VQY9 A0A1J1IZY5 D6W9N6 A0A096XDB4 Q171V8 A0A336LJV2 A4KWW3 A4ZKB7 H9JAC5 Q8WSZ4 Q8WQP9 A4ZKB8 A0A1B0CZP4 Q14TE5 B0WAJ2 A0A1B0CZP1 T1IDE9 A0A1Y1LII3 A0A1W4WVL1 A0A182PS55 A0A2S2QMZ6 W5JSC3 A0A023EUP4 A0A3M6TZ36 N6UAX5 A0A023ERW8 A0A0F8B2S9 A0A1J1J657 T1PQM9 E2BGQ2 A0A336MAE4 A0A3B4Y3V9 A0A3B4U151 A0A1S4EFC2 Q173B1 A0A0K8S5D1 A0A182IPM3 A0A182TJZ6 A0A023ERW2 A0A1S3D6B4 A0A1S3D7T3 A0A0P6J0V5 A0A0A9XP67 A0A1S4G6X1 A0A2S2QTL0 A0A1S4ECH4 M1FXD6 A0A182R814 T1I119 U5EY18 A0A182T7Z9 A0A2D4KYW2 A0A2S2QIW8 A0A1B0DIJ1 A0A182KWY6 A0A182HJS3 Q7QDJ5 A0A182XK65 A0A182V4X1
PDB
4YMK
E-value=9.42805e-34,
Score=352
Ontologies
GO
PANTHER
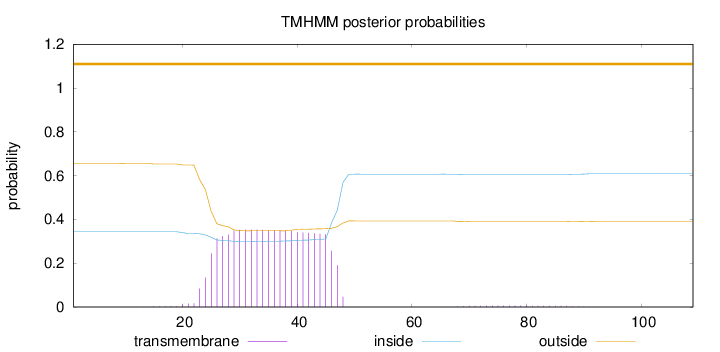
Topology
Length:
109
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.99448
Exp number, first 60 AAs:
7.87402
Total prob of N-in:
0.34395
outside
1 - 109
Population Genetic Test Statistics
Pi
217.928957
Theta
160.443255
Tajima's D
1.126693
CLR
0
CSRT
0.693165341732913
Interpretation
Uncertain
