Gene
KWMTBOMO03328
Annotation
serine_protease_like_protein_[Bombyx_mandarina]
Location in the cell
Extracellular Reliability : 1.18 PlasmaMembrane Reliability : 1.487
Sequence
CDS
ATGAGAAGTATATTGAGTCTTACTGTTGTACTATTTTGTTTGGGTTTTCAGGTTGCGTTAGCGCAGACACATGGATGCAAATGTGGTAAGGCTTCGGGTTCCATAGTAGAAATGAGGATTGTCGGCGGCCGAAGAGCGGTTCCACACTCCTTTCCGTGGACAGTGGCGATCCTGAAGCAAAAGTTGCTGCACTGCGGTGGCGCTCTCATCACAAATGAGCATGTACTCAGCGCGGGCCATTGCTTCAAATGGGATGAACCCAAAATTATGAGGGTTTTGTTAGGACTGGATCATTTGGACAATATGACCGGAGTAGAAATTCGCACGATATCGAACGTGAAGATTCACGAACACTTTACTTCGACAGCTCTACGAGATGAGCACGATATCGCCATAGTAACATTGAATAAACCAGTTTTTTTCGGAGATAACATTATACCGATATGCTTGCCGAGCCCAGGTGCAGATTTTGCGAATAGAATGGGTACAATAGTGGGCTGGGGTCGGGTGGGTGTGGACAAGTCATCCTCGAGAACCCTTTTAAAGGCCTCACTACGGATACTTTCACAAGAACAATGCATGAAATCGGAATTGAAACAACATCTCAAACCAACGATGATGTGCGCTTTCAGTAAAGGAATAGACGGTTGCCAGGGAGACAGTGGCGGTCCGCTAGTAGTGTTAGAGCCCACAGAACGATACGTGCAAGCGGGAATAGTTTCTTGGGGCATAGGATGCGCTGATCCAAGATATCCAGGTATATAA
Protein
MRSILSLTVVLFCLGFQVALAQTHGCKCGKASGSIVEMRIVGGRRAVPHSFPWTVAILKQKLLHCGGALITNEHVLSAGHCFKWDEPKIMRVLLGLDHLDNMTGVEIRTISNVKIHEHFTSTALRDEHDIAIVTLNKPVFFGDNIIPICLPSPGADFANRMGTIVGWGRVGVDKSSSRTLLKASLRILSQEQCMKSELKQHLKPTMMCAFSKGIDGCQGDSGGPLVVLEPTERYVQAGIVSWGIGCADPRYPGI
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
I4DDG2
B9X253
F8WS77
G5EMP4
G5EMP3
I4DDG3
+ More
B9X2I4 I4DDG4 A0A097ZMX2 I7H7W0 I7H0C7 G5EMP2 A0A212F4K4 D9HQ64 A0A2H1V284 D9HQ22 A0A194Q376 A0A194RA86 G9D495 G9D4A6 G9D496 G9D4A5 G9D4A3 G9D4A4 G9D4A2 G9D493 E9IHW6 A0A0L7L5B1 F4X2V5 A0A195D4W1 E2BDT3 A0A195EIX8 G5EMR5 K7IT89 A0A195F5W7 I7GYE3 E5KHE3 Q0Q605 D0EXD7 D9HQ87 A0A026WA15 H2D0L3 H2D0L2 H2D0L4 H2D0L1 A0A2A4IY89 A0A220K8P6 E2AFZ1 A0A158P3H0 A0A3L8E3C9 A0A0C9RD19 A0A0C9QMS3 A0A026WAC6 A0A0C9R9I2 A0A195EK23 F4X2V2 A0A195BW73 A0A3L8E3W1 A0A151WZG8 A0A068F4F5 A0A087ZRJ0 B3GC92 B3GC76 A0A1V1FVE0 A0A3Q0IU66 E2B8I2 A0A3P9L447 A0A3P9MER2 A0A2A3E7J8 A0A3P9L477 A0A151IF83 A0A068FBA1 A0A0K2UHZ0 E2AFY8 A0A2J7RLX2 A0A3P9MF96 E0VPJ3 A0A1A9WE67 A0A336M269 K7J296 A0A3P9L3W6 A0A3L8DR71 A0A026WDZ3 A0A1V1FVC3 U4U2H2 B4LD08 A0A0K2TQY0 E2AVK1 A0A067QVB6 A0A1B6BZ17 A0A3P9MDC2 A0A0N0BDH6 E0VW10 A0A3P9MF45 A0A139WME7 A0A0J7KZW0 D6W9P0 A0A1B6MS67 A0A151WUF5
B9X2I4 I4DDG4 A0A097ZMX2 I7H7W0 I7H0C7 G5EMP2 A0A212F4K4 D9HQ64 A0A2H1V284 D9HQ22 A0A194Q376 A0A194RA86 G9D495 G9D4A6 G9D496 G9D4A5 G9D4A3 G9D4A4 G9D4A2 G9D493 E9IHW6 A0A0L7L5B1 F4X2V5 A0A195D4W1 E2BDT3 A0A195EIX8 G5EMR5 K7IT89 A0A195F5W7 I7GYE3 E5KHE3 Q0Q605 D0EXD7 D9HQ87 A0A026WA15 H2D0L3 H2D0L2 H2D0L4 H2D0L1 A0A2A4IY89 A0A220K8P6 E2AFZ1 A0A158P3H0 A0A3L8E3C9 A0A0C9RD19 A0A0C9QMS3 A0A026WAC6 A0A0C9R9I2 A0A195EK23 F4X2V2 A0A195BW73 A0A3L8E3W1 A0A151WZG8 A0A068F4F5 A0A087ZRJ0 B3GC92 B3GC76 A0A1V1FVE0 A0A3Q0IU66 E2B8I2 A0A3P9L447 A0A3P9MER2 A0A2A3E7J8 A0A3P9L477 A0A151IF83 A0A068FBA1 A0A0K2UHZ0 E2AFY8 A0A2J7RLX2 A0A3P9MF96 E0VPJ3 A0A1A9WE67 A0A336M269 K7J296 A0A3P9L3W6 A0A3L8DR71 A0A026WDZ3 A0A1V1FVC3 U4U2H2 B4LD08 A0A0K2TQY0 E2AVK1 A0A067QVB6 A0A1B6BZ17 A0A3P9MDC2 A0A0N0BDH6 E0VW10 A0A3P9MF45 A0A139WME7 A0A0J7KZW0 D6W9P0 A0A1B6MS67 A0A151WUF5
Pubmed
EMBL
AB684322
BAM15952.1
AB485779
BAH23567.1
AB645766
BAK52270.1
+ More
AB678223 BAL04890.1 AB678222 BAL04889.1 AB684323 BAM15953.1 AB489911 BAH24266.1 AB684324 BAM15954.1 AB924381 BAP76070.1 AB738397 BAM34530.1 AB738398 BAM34531.1 AB678221 BAL04888.1 AGBW02010347 OWR48666.1 HM023819 JF777147 ADJ58552.1 AEU11492.1 ODYU01000347 SOQ34950.1 HM023777 JF777154 ADJ58510.1 AEU11499.1 KQ459551 KPI99991.1 KQ460473 KPJ14532.1 JF777148 AEU11493.1 JF777159 AEU11504.1 JF777149 AEU11494.1 JF777158 AEU11503.1 JF777156 AEU11501.1 JF777157 AEU11502.1 JF777155 AEU11500.1 JF777146 AEU11491.1 GL763294 EFZ19888.1 JTDY01002816 KOB70663.1 GL888593 EGI59210.1 KQ976842 KYN07912.1 GL447689 EFN86139.1 KQ978801 KYN28215.1 AB678438 BAL14423.1 AAZX01003313 KQ981805 KYN35464.1 AB734410 BAM29297.1 HQ130738 ADQ43543.1 DQ520139 ABG75840.1 GQ911573 ACX54054.1 HM023842 ADJ58575.1 KK107304 EZA52937.1 JN393685 AEX97001.1 JN393684 AEX97000.1 JN393686 AEX97002.1 JN393683 AEX96999.1 NWSH01004663 PCG64795.1 MF319737 ASJ26451.1 GL439184 EFN67641.1 ADTU01007955 ADTU01007956 QOIP01000001 RLU27156.1 GBYB01004791 JAG74558.1 GBYB01004789 JAG74556.1 EZA52933.1 GBYB01004790 JAG74557.1 KYN28214.1 EGI59207.1 KQ976403 KYM92218.1 RLU27153.1 KQ982649 KYQ53176.1 KJ512103 AID60326.1 EU669773 EU669774 EU669775 EU669776 EU669777 EU669778 EU669779 EU669780 EU669781 EU669782 EU669785 EU669786 ACD69531.1 ACD69532.1 ACD69533.1 ACD69534.1 ACD69535.1 ACD69536.1 ACD69537.1 ACD69538.1 ACD69539.1 ACD69540.1 ACD69543.1 ACD69544.1 EU669757 EU669758 EU669759 EU669760 EU669761 EU669762 EU669763 EU669764 EU669765 EU669766 EU669767 EU669768 EU669769 EU669770 EU669771 EU669772 ACD69515.1 ACD69516.1 ACD69517.1 ACD69518.1 ACD69519.1 ACD69520.1 ACD69521.1 ACD69522.1 ACD69523.1 ACD69524.1 ACD69525.1 ACD69526.1 ACD69527.1 ACD69528.1 ACD69529.1 ACD69530.1 FX985325 BAX07338.1 GL446332 EFN88016.1 KZ288360 PBC27021.1 KQ977800 KYM99571.1 KJ512116 AID60339.1 HACA01019950 CDW37311.1 EFN67638.1 NEVH01002683 PNF41809.1 DS235366 EEB15299.1 UFQT01000349 SSX23441.1 AAZX01018692 QOIP01000005 RLU22796.1 KK107293 EZA53249.1 FX985305 BAX07318.1 KB631815 ERL86503.1 CH940647 EDW69889.1 HACA01011073 CDW28434.1 GL443122 EFN62552.1 KK853216 KDR09771.1 GEDC01030802 JAS06496.1 KQ435859 KOX70461.1 DS235816 EEB17566.1 KQ971312 KYB29219.1 LBMM01001704 KMQ95868.1 EEZ99213.1 GEBQ01001197 JAT38780.1 KQ982736 KYQ51468.1
AB678223 BAL04890.1 AB678222 BAL04889.1 AB684323 BAM15953.1 AB489911 BAH24266.1 AB684324 BAM15954.1 AB924381 BAP76070.1 AB738397 BAM34530.1 AB738398 BAM34531.1 AB678221 BAL04888.1 AGBW02010347 OWR48666.1 HM023819 JF777147 ADJ58552.1 AEU11492.1 ODYU01000347 SOQ34950.1 HM023777 JF777154 ADJ58510.1 AEU11499.1 KQ459551 KPI99991.1 KQ460473 KPJ14532.1 JF777148 AEU11493.1 JF777159 AEU11504.1 JF777149 AEU11494.1 JF777158 AEU11503.1 JF777156 AEU11501.1 JF777157 AEU11502.1 JF777155 AEU11500.1 JF777146 AEU11491.1 GL763294 EFZ19888.1 JTDY01002816 KOB70663.1 GL888593 EGI59210.1 KQ976842 KYN07912.1 GL447689 EFN86139.1 KQ978801 KYN28215.1 AB678438 BAL14423.1 AAZX01003313 KQ981805 KYN35464.1 AB734410 BAM29297.1 HQ130738 ADQ43543.1 DQ520139 ABG75840.1 GQ911573 ACX54054.1 HM023842 ADJ58575.1 KK107304 EZA52937.1 JN393685 AEX97001.1 JN393684 AEX97000.1 JN393686 AEX97002.1 JN393683 AEX96999.1 NWSH01004663 PCG64795.1 MF319737 ASJ26451.1 GL439184 EFN67641.1 ADTU01007955 ADTU01007956 QOIP01000001 RLU27156.1 GBYB01004791 JAG74558.1 GBYB01004789 JAG74556.1 EZA52933.1 GBYB01004790 JAG74557.1 KYN28214.1 EGI59207.1 KQ976403 KYM92218.1 RLU27153.1 KQ982649 KYQ53176.1 KJ512103 AID60326.1 EU669773 EU669774 EU669775 EU669776 EU669777 EU669778 EU669779 EU669780 EU669781 EU669782 EU669785 EU669786 ACD69531.1 ACD69532.1 ACD69533.1 ACD69534.1 ACD69535.1 ACD69536.1 ACD69537.1 ACD69538.1 ACD69539.1 ACD69540.1 ACD69543.1 ACD69544.1 EU669757 EU669758 EU669759 EU669760 EU669761 EU669762 EU669763 EU669764 EU669765 EU669766 EU669767 EU669768 EU669769 EU669770 EU669771 EU669772 ACD69515.1 ACD69516.1 ACD69517.1 ACD69518.1 ACD69519.1 ACD69520.1 ACD69521.1 ACD69522.1 ACD69523.1 ACD69524.1 ACD69525.1 ACD69526.1 ACD69527.1 ACD69528.1 ACD69529.1 ACD69530.1 FX985325 BAX07338.1 GL446332 EFN88016.1 KZ288360 PBC27021.1 KQ977800 KYM99571.1 KJ512116 AID60339.1 HACA01019950 CDW37311.1 EFN67638.1 NEVH01002683 PNF41809.1 DS235366 EEB15299.1 UFQT01000349 SSX23441.1 AAZX01018692 QOIP01000005 RLU22796.1 KK107293 EZA53249.1 FX985305 BAX07318.1 KB631815 ERL86503.1 CH940647 EDW69889.1 HACA01011073 CDW28434.1 GL443122 EFN62552.1 KK853216 KDR09771.1 GEDC01030802 JAS06496.1 KQ435859 KOX70461.1 DS235816 EEB17566.1 KQ971312 KYB29219.1 LBMM01001704 KMQ95868.1 EEZ99213.1 GEBQ01001197 JAT38780.1 KQ982736 KYQ51468.1
Proteomes
UP000007151
UP000053268
UP000053240
UP000037510
UP000007755
UP000078542
+ More
UP000008237 UP000078492 UP000002358 UP000078541 UP000053097 UP000218220 UP000000311 UP000005205 UP000279307 UP000078540 UP000075809 UP000005203 UP000079169 UP000265180 UP000242457 UP000235965 UP000009046 UP000091820 UP000030742 UP000008792 UP000027135 UP000053105 UP000007266 UP000036403
UP000008237 UP000078492 UP000002358 UP000078541 UP000053097 UP000218220 UP000000311 UP000005205 UP000279307 UP000078540 UP000075809 UP000005203 UP000079169 UP000265180 UP000242457 UP000235965 UP000009046 UP000091820 UP000030742 UP000008792 UP000027135 UP000053105 UP000007266 UP000036403
Interpro
Gene 3D
ProteinModelPortal
I4DDG2
B9X253
F8WS77
G5EMP4
G5EMP3
I4DDG3
+ More
B9X2I4 I4DDG4 A0A097ZMX2 I7H7W0 I7H0C7 G5EMP2 A0A212F4K4 D9HQ64 A0A2H1V284 D9HQ22 A0A194Q376 A0A194RA86 G9D495 G9D4A6 G9D496 G9D4A5 G9D4A3 G9D4A4 G9D4A2 G9D493 E9IHW6 A0A0L7L5B1 F4X2V5 A0A195D4W1 E2BDT3 A0A195EIX8 G5EMR5 K7IT89 A0A195F5W7 I7GYE3 E5KHE3 Q0Q605 D0EXD7 D9HQ87 A0A026WA15 H2D0L3 H2D0L2 H2D0L4 H2D0L1 A0A2A4IY89 A0A220K8P6 E2AFZ1 A0A158P3H0 A0A3L8E3C9 A0A0C9RD19 A0A0C9QMS3 A0A026WAC6 A0A0C9R9I2 A0A195EK23 F4X2V2 A0A195BW73 A0A3L8E3W1 A0A151WZG8 A0A068F4F5 A0A087ZRJ0 B3GC92 B3GC76 A0A1V1FVE0 A0A3Q0IU66 E2B8I2 A0A3P9L447 A0A3P9MER2 A0A2A3E7J8 A0A3P9L477 A0A151IF83 A0A068FBA1 A0A0K2UHZ0 E2AFY8 A0A2J7RLX2 A0A3P9MF96 E0VPJ3 A0A1A9WE67 A0A336M269 K7J296 A0A3P9L3W6 A0A3L8DR71 A0A026WDZ3 A0A1V1FVC3 U4U2H2 B4LD08 A0A0K2TQY0 E2AVK1 A0A067QVB6 A0A1B6BZ17 A0A3P9MDC2 A0A0N0BDH6 E0VW10 A0A3P9MF45 A0A139WME7 A0A0J7KZW0 D6W9P0 A0A1B6MS67 A0A151WUF5
B9X2I4 I4DDG4 A0A097ZMX2 I7H7W0 I7H0C7 G5EMP2 A0A212F4K4 D9HQ64 A0A2H1V284 D9HQ22 A0A194Q376 A0A194RA86 G9D495 G9D4A6 G9D496 G9D4A5 G9D4A3 G9D4A4 G9D4A2 G9D493 E9IHW6 A0A0L7L5B1 F4X2V5 A0A195D4W1 E2BDT3 A0A195EIX8 G5EMR5 K7IT89 A0A195F5W7 I7GYE3 E5KHE3 Q0Q605 D0EXD7 D9HQ87 A0A026WA15 H2D0L3 H2D0L2 H2D0L4 H2D0L1 A0A2A4IY89 A0A220K8P6 E2AFZ1 A0A158P3H0 A0A3L8E3C9 A0A0C9RD19 A0A0C9QMS3 A0A026WAC6 A0A0C9R9I2 A0A195EK23 F4X2V2 A0A195BW73 A0A3L8E3W1 A0A151WZG8 A0A068F4F5 A0A087ZRJ0 B3GC92 B3GC76 A0A1V1FVE0 A0A3Q0IU66 E2B8I2 A0A3P9L447 A0A3P9MER2 A0A2A3E7J8 A0A3P9L477 A0A151IF83 A0A068FBA1 A0A0K2UHZ0 E2AFY8 A0A2J7RLX2 A0A3P9MF96 E0VPJ3 A0A1A9WE67 A0A336M269 K7J296 A0A3P9L3W6 A0A3L8DR71 A0A026WDZ3 A0A1V1FVC3 U4U2H2 B4LD08 A0A0K2TQY0 E2AVK1 A0A067QVB6 A0A1B6BZ17 A0A3P9MDC2 A0A0N0BDH6 E0VW10 A0A3P9MF45 A0A139WME7 A0A0J7KZW0 D6W9P0 A0A1B6MS67 A0A151WUF5
PDB
4DUU
E-value=1.54257e-31,
Score=338
Ontologies
GO
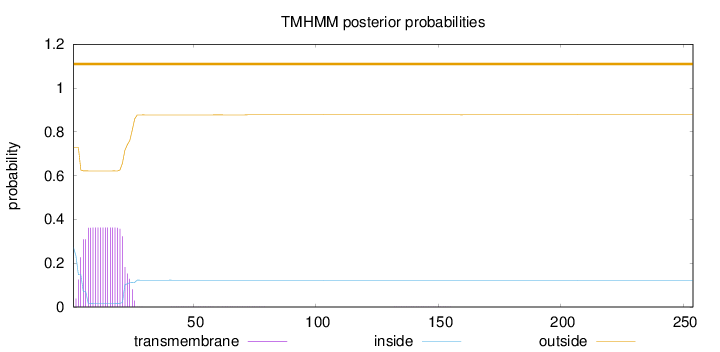
Topology
SignalP
Position: 1 - 21,
Likelihood: 0.995687
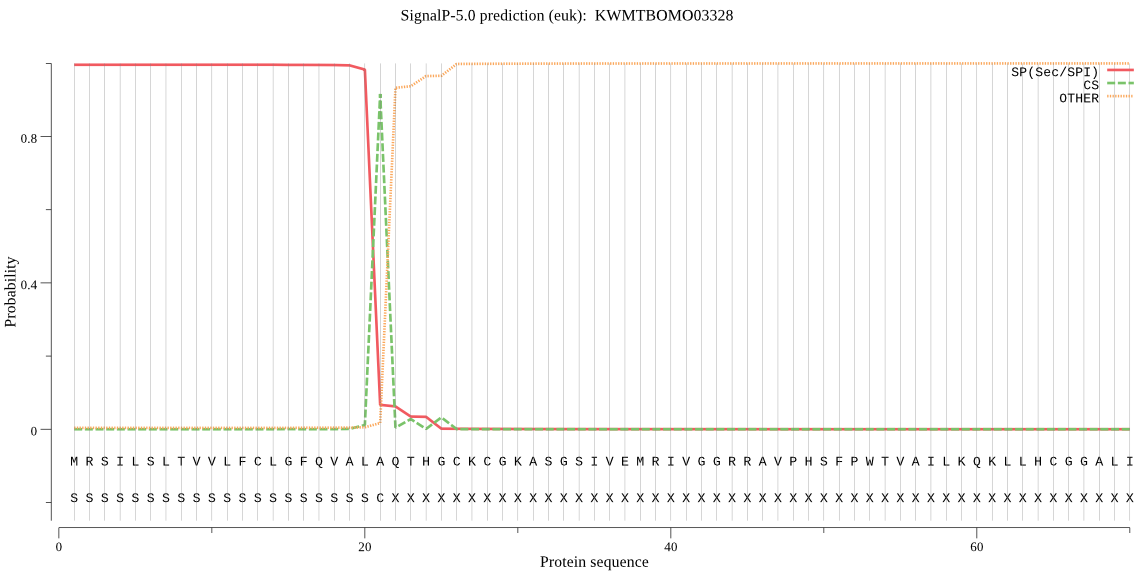
Length:
254
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.98617
Exp number, first 60 AAs:
6.97672
Total prob of N-in:
0.27205
outside
1 - 254
Population Genetic Test Statistics
Pi
195.585762
Theta
151.528185
Tajima's D
0.549278
CLR
1.298744
CSRT
0.531873406329683
Interpretation
Uncertain
