Gene
KWMTBOMO03326
Pre Gene Modal
BGIBMGA013209
Annotation
PREDICTED:_dynamin-like_120_kDa_protein?_mitochondrial_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.474
Sequence
CDS
ATGATTAACGAGCAGTATCAGCGACTAGTAGAACAAGTAGGCATTCTTCAAGAAGGGGAGGAAGCTGGTGCCCGTTCCCGGACGCCAGCAGGAGTGAAGTCAAAACAAATAAATGAAGAAACAAGAACCAGTGGGCAAAGCCAACAGGTTGAACAAATGAAAAGAAGCCAAGGCACAAACAGTAAATTGGATGATTTCTTAAAAAGACAATATGCTAATTTTAAAGCCTTTATACGAACAACATCAAACATAGACGTAGATTTGATTGCTGATAAATGGGAATTTGAAGACTCTTTAAAAACACTACACGAACGATGGGCTGCTATCGATAGCCTTCACTGGGAAATCGAATCCGAGATGGACGGAGAAAATGAAACATACGCCGCCATGTACGATAGATACGAGAAGAAATTTAATACTCTCAAGAAAACATTGAACACAAAGATGTGGTCGGTAGCTCATAGATCGAATGCTACTCCACAATTGGAAATACCTGTCTTCAGCGGAAATTATAATCAGTGGGTTTCTTTCAAAGATTTATTCACGGAAGCTGTTCACAACAATCCATCTCTATCTAACAGTCAAAAAATGCAACACCTGAAAACTCCGGTGTCAACTGAACAAAACATTGCGAAACAACGCATAACACGAAATGAGAATGTTTCAAAAACCTCTAATGCTTCGCTAAATGAGAGTTCAGAAGTTTTATTAGCTACTGCTTTGGTGAAGGTGAAAGCGGCTGATGGAACCTATTACAACATGCGAGCTTTGTTAGATCAAGGCTCGCAAATATCTTTGATAACTGAAGCAGCAGCCCAAACATTAGGATTAAAGCGTCAAAGATGCAAAGGGGTGATATTTGGTGTAGGCCAAAACGAAAACAGCTGCAAAGGAAAGGTAATCATAGAATGCTCCTCAATTAACAATGACTTCAATTTCGAACTAGAGGCACTCATTATGAACAATTTAATAAAAAGTTTACCAAACAAAACTTTTTCTAAGCCAGCGTGGTCATATTTGAATAATATCAATTTAGCTGATCCCGAGTTTTACAAGAGTCGGCCAGTCGATCTACTGTTCGGTGCGGATATCTACGCTAACATTATTCTGGGAGGAGTGATTAAAGGAGAAACCTTACAACAACCCATGGCTCAACAATCACAGTTAGGATGGTTTTTATGCGGAAGTATTAAAACTTATCATTGTAATGTGGCTCTCAATAACGTGGAAGATATACATAAATTTTGGGAGGTGGAAGAAATCAACGAATCGACGAATATTTCCGTAGAAGATCAGGAATGTATCGACTTTTACTGCTCAACAACAAAACGCATGGAAAATGGTCGATATGAAGTGAGATTGCCTCTAAAGCAAGATTTAAAACATAATTTAGGAATGAAGGCATTCGAAAAGGCGACAACACATTTAGATAAATCATGCAAAGTCCTGGAAGATGGTCAGTGTAGCATAATAGTCGATCAATTGATCCACGAATACAAAGAATTAACTTACTATAATGAGCTACTTATGAATCAACATTTGAATGAGCACTACAGACAGCGTCGTGGTTTTATCAACGGTGTCGGATATCTTGCTAACTCTCTGTTCGGAGTATTAGATGAACATTTCGCAGAAAAATACTCGCAAGACATAGAGCTCGTAAAACATAATGAACAGTACTTAGTGAGTTTATGGAAAAACCAAACTTCGATCGTCGAATCTGAATACAACTTACTAAAAAGAACAGAAGAAGTCATGTCTAAGCAACATAAGACTATTAATAAACACATAACTAACCTAGAAAACATGTCAAATCGACTTCAACAGCAAGTAAACAACATATCGATTCTACAAGAATTTACCTTAATGTCCATTATTACAACTAATTTATTGCACAATTTGAAAGGGTCGCAAAATATATTGTTAGACATTTTAACTAACATACATCATGGTCAATTCAACATTCATTTACTTACTCCAGAACAACTTCAAAAGGAACTAAACATTATCACCGGATATTTACCGGAAGACACAATTCTTCCCGTAGAACCGCAGAATATTAAGAATTTGTATCCTCTGTTAAACATAAAGGCTCGTATTACTCTACAATATTTCTTATTCGAGATAACATTTCCACTCATTTATCAAGATCGATTCAAACTTTATAAAGTACATTCTATTCCTCATCAAAGCGGAAATATGACTATTGAAGTAGTTCCGGCATACGAATACATTGCTACCGACTTGAAAAGAGATTCCTTTCTTGAACTCACCACCGAAGAAGTCTTATCCTGTAATCAACAAGAAGAAGCTTACTACTGCAAGCTTAGAACGCCGATTCTTCATTTACGACCGGAGACTAAGTTTTGTAAGATTAAACTTAATAGCAACGTATGCAAATTAAAGTATTCTAATTGTGAAACCAAAATAATAGCACTAACTCATCTCTCTTTATATTTATATCACTGCTGTGGCACCTGCGAAGTAAAGGTCATGTGCGGAGAACATATTGCCTTGAAACAGTTGAATAACACTGGTCGTCTGTATCTAGAGCCAGGTTGCCTCATAAAGGGATCAGATTATACACTTCATATCCCTAAAACAAACAACAATAAGCTGGAGATTCAGTCTAACATTTACGTTCCAATAATAGATCCCATAAATAATATCATAAACAGCTCTCTTCTCAACACTGAGAACCTGAATGTCAGCGAGTCTAAAGATTTGCAGAACGCTCTCCAGAAACTACACGTGCGTATTGAAGATCTAGAAAACTCCCCGCCAGTATTGGAGACTTCATTAACATCTCACGATATTCATCAGTATGTGATAACATACATCATCGTCATCGTCATCGTGCTGGCCACCGGCGTGCTACTCTGGCGACGACGACGGTTGAGTGCGAACCGGGCGATCATCTCTGACGGCATCGAGCTTCAACAGTATAGTGAATGTGAAAATGTAAATAGTGCTAAGGCAGGGTTTAGTGCGCGACCTTCTGTGGTGGACCCTTCGCCTGGTGATGTGAACATTTATGCCAAAGTGTCAAACAAAGCGTGCTCTCCAATACCAAGAAAACCTATATGGACCCTAAGTTCTTAA
Protein
MINEQYQRLVEQVGILQEGEEAGARSRTPAGVKSKQINEETRTSGQSQQVEQMKRSQGTNSKLDDFLKRQYANFKAFIRTTSNIDVDLIADKWEFEDSLKTLHERWAAIDSLHWEIESEMDGENETYAAMYDRYEKKFNTLKKTLNTKMWSVAHRSNATPQLEIPVFSGNYNQWVSFKDLFTEAVHNNPSLSNSQKMQHLKTPVSTEQNIAKQRITRNENVSKTSNASLNESSEVLLATALVKVKAADGTYYNMRALLDQGSQISLITEAAAQTLGLKRQRCKGVIFGVGQNENSCKGKVIIECSSINNDFNFELEALIMNNLIKSLPNKTFSKPAWSYLNNINLADPEFYKSRPVDLLFGADIYANIILGGVIKGETLQQPMAQQSQLGWFLCGSIKTYHCNVALNNVEDIHKFWEVEEINESTNISVEDQECIDFYCSTTKRMENGRYEVRLPLKQDLKHNLGMKAFEKATTHLDKSCKVLEDGQCSIIVDQLIHEYKELTYYNELLMNQHLNEHYRQRRGFINGVGYLANSLFGVLDEHFAEKYSQDIELVKHNEQYLVSLWKNQTSIVESEYNLLKRTEEVMSKQHKTINKHITNLENMSNRLQQQVNNISILQEFTLMSIITTNLLHNLKGSQNILLDILTNIHHGQFNIHLLTPEQLQKELNIITGYLPEDTILPVEPQNIKNLYPLLNIKARITLQYFLFEITFPLIYQDRFKLYKVHSIPHQSGNMTIEVVPAYEYIATDLKRDSFLELTTEEVLSCNQQEEAYYCKLRTPILHLRPETKFCKIKLNSNVCKLKYSNCETKIIALTHLSLYLYHCCGTCEVKVMCGEHIALKQLNNTGRLYLEPGCLIKGSDYTLHIPKTNNNKLEIQSNIYVPIIDPINNIINSSLLNTENLNVSESKDLQNALQKLHVRIEDLENSPPVLETSLTSHDIHQYVITYIIVIVIVLATGVLLWRRRRLSANRAIISDGIELQQYSECENVNSAKAGFSARPSVVDPSPGDVNIYAKVSNKACSPIPRKPIWTLSS
Summary
Uniprot
ProteinModelPortal
Ontologies
GO
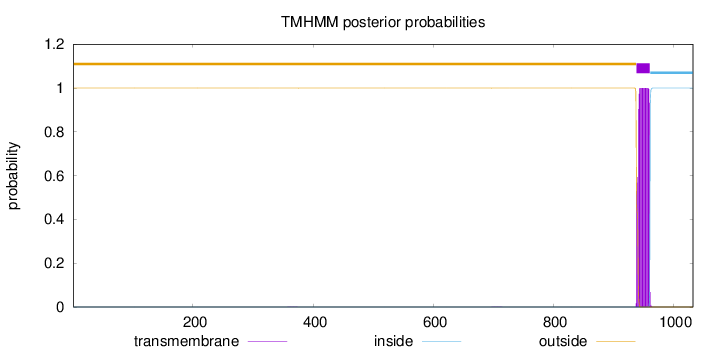
Topology
Length:
1033
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.60058
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00023
outside
1 - 938
TMhelix
939 - 961
inside
962 - 1033
Population Genetic Test Statistics
Pi
396.005246
Theta
209.331736
Tajima's D
1.964368
CLR
113.987336
CSRT
0.87835608219589
Interpretation
Uncertain
