Gene
KWMTBOMO03320
Pre Gene Modal
BGIBMGA006467
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_chaoptin_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.088 Nuclear Reliability : 1.601 PlasmaMembrane Reliability : 1.508
Sequence
CDS
ATGTTGCTGTTGCTACTTCTATGTTTACTTCCGTACATCGCCACCCAACAACAATGGGTGCCTTGTTCAGAGTTGAACGACGACTTACGCTACCCCTGTCGGTGTCGAGTGCAAGTTGACAGAGCCCTCCAGTTGAGAATCTTGATGAATTGTGACCGAGTGGTCTTTGCTGGAGACTTCCCGACTTTGCCATATGGCGCTCCTATTATTTCCTTCAGTCAACGTTGGGCTGGACAACAGACGCTGCCGATGCAGATATTCTCCACGTACGGGCTGCCTTTACGCGAGCTAGATTTCTCTCACAACAGTCTACGACGCTTGCCAGATCGTCTCCTAGCTGCCGTCCGGGGAAACCTGACCAAGTTGGCATTAGCTGATAATCTACTCGGTGACAACTTGAATCCTATATTCTCGACTGCAGAACTCCACAATTTGCCCGCTTTAGAAGAATTGGATCTTAGCGGCAATAGCATACGAGGGATCGAAGAGGGCTTGTTAATCGGATGTGATGTTCTACAGGTACTCCGCCTTGATCGTAACAACATAAACTTCATTCCGTCCTCTTCGCTGAACGGGCCACAATCATTGAGAGTCCTTTCGCTTCAGGAAAATAAAATAGCTTTGTTGCGTCAAGCATCGTTTGTTTCACAAAAGTCATTAGAAGAGATTAACTTACATGGAAACATGATATCGACTATTGAAGGAGGGGCATTTCTAGGCTTAAAAGACCTGCAAATTCTAGATCTAGGCCGCAACAGACTGTCTAAGTTCAATAGTGACGTGTTTCAAGGCGCTGAAAACTTGGAGAAGCTGGACTTGTCTGAGAACTTTATAAACGATTTCCCCACCGTGGCCATCAAAACGTTTGGCGGCTTAAGTCATTTGAATTTGTCTAGTAATATGATTTCCAACATCGACAATAGCCACCTAAATTCGCTGTCATCGTTACAAGTTTTGGACTTAAGCCGGAATAATTTGGTTAAGCTCACCCCGGGAACATTTGTAGGACTGACTGAATTGAGTTACTTGGACATCGGAGTGAACTCTTTACGAACTGTCGAAGATAATGCCTTCGATGGACTGACAAGTCTTAAAACTCTTCTCTTGCGAGACAACAACATTCTGCTGATCCCTGCTACTGCACTGTCACGTTTGCCAAATCTAGTTTCAGTGCATCTAGGATTTAACAGAGTAACGGCTCTATCAAACGATATACTTAGATCGGTTTCAGATAGAGTGACTTCTTTAGTTTTGTCAAGAAATGTTATACGAGAACTACCGACGGCAGCATTTGATCATTTCAAAATGTTAAGGCACTTGGATTTGTCAGGAAATTTACTGAATACGATTGCTGCTGAAGTATTCGATGGATTGGAACAATCGCTACAGTTTCTGTCTTTAAGCCAAAACAGAATTCTAGGATTCACTGGACAACAACTAAAGTTTGTGAGCCTTTGGTTTCTAGATATTTCTGGCAATCAGATATCAAATGTACCTGTCGAGGCTTTTGAGTCTATACCGAGTTTGACACATTTAAATATGAGCCGTAATCAGCATATTAACGTTTTACCTCAAAACTTATTCCAACACAATCAAGCATTATTGACTATTGATTTAAGCTATAATGGATTAAAAGCTCTCCCCGTGAATTTATTCTCCAAAAATTTTAACTTACAGCAAATACATTTATCAAATAATCTTCTGCAAGAAATTTCTGAGAACACCTTTAAGAATCTTCGAAATTTGACACATCTAGATCTGTCATATAATAATATTGTCAATTTAAGAACACCATCGTTTGTCAATGTGATGTCTATACAATATTTGTCACTCAAAGGTAATCAGTTAAGCGCTTTTAAGGGAGAATTTTTCAACACAGGAACAAGTCTAGAAGTAATCGATCTTTCAGACAATCAGCTTAGCTACTTGTTTCCCTCTGCCTTCAAAATACACCCTCGTTTGCGAGAAATCAAACTTGCAAACAATAAGTTCAACTTTTTCCCGTCGGAACTTATAAGTACTTTGCAGTATTTAGAATTAGTTGATTTATCTGGAAACGCTTTGAAAAATGTCGATGAGCTTGATTTTGCTCGCCTTCCAAAACTGAGGACAATTTTACTAGCAAGAAATGAATTGGAAACAGTTAGCGAAATGGCTTTCCATAACTCGTCGCAAATACAACAATTAGATTTGTCTCAGAATAAAATCGACCGCTTGGGAGATCGACTTTTTGAAGGACTAATAAGATTAGAAACTTTAAACTTAGCAGGGAATATGCTTGCTGAACTACCTGATAGTATTTTTGAAAGGAATCGTTTACATATGCTCGAAAATATAAATCTAAGTGGAAATTTGTTCGAACATCCACCCCTGAAAGCTTTACAGAAGCAATACTTCTTTGTTTCTGCTGTAGATTTGTCACACAACAAAATAGTTGACATACCAGCCGAAGACAGTGTTTTGAGACTAAACATTTCTAATAACCCCATAGTCATGATCATGGACGGAAATTTTGAGGGTCTAACGTCGCTGCGTGTGTTAAATATGAATAATCTAGACAAATGCACAAGAATTGAAAAAAATGCCTTTAGATCTCTTCCAAACCTCATTGAATTAAGGGCTTTTGGCTATCCAAGACTTGGCTATTTTGATGTTCAAGGCACTTTGCAATATTTATTTGCACTAGAAAAATTAGATGTTGAACTGAAAGACACAAATATTGGGCCAGATCAGCTACACTCTACGTTACATCCTCGTCTCGAAGAATTGGGCATTAGAGGATCTCGATTAAAAACAGTGTCTTCGGGTGTGTTAGCTGGTTTAAAAGCTCCAGCTATAGTAGTTAGACTCCGTAATACTTCTGTATCAACATTACCACCTGCTTTGCTATTTCCTCTACCGAGGTCATCGCAAATAACAATTGATGTTGGCGGCAGTCAATTAGAGACTTTACAACCTCAGTTGCTGGTGGCACTCGACGATCGACGTGCAGATCTATCGATGTTGGGTTTGGATACCAATCCTATCAGATGTGACTGCAATGCTCGGGCTTTGAGACGATGGCTACCAAATGTTGGAATTGATAATATTCGCTGTCAGTCACCAGACTATTTATCTGGATATTTACTTGTTGAAATAGGAGACGATGAACTAACTTGTGACTCAAAACGCCGAACAACAGCCACATCAACTTCAAGTGTTTCAACAACATCTCCACCACGTTTAGTTCACCGCACATCTTCTGAACCAGATATTATATGGTCAGTAGCTCCATCTCACGATAGACCAAAGATATCCGGAGAACCGAAAGGCGCTCCTGTTATCGGTGTTGCAACGGCGTCGAACGATGACAATCTCATAATTGGAATAGTTAGTGGTGTCGTTGCTTTTATAGCTATTTTAATAGTAGCCATTTGTATTATAAGATTAAAGATGACTACAACATCATACAGAGGTGGACCACTAGCTAATAGCCCAACAGGCGGTGCGCCAGCAGTATGGGGTCCACCTTGGCCCGGTTATACCGGCACAATGCCACCAGCCTCTCTATCAACAGCAACATTGCCTCATAAGGTTCAACCAGGACCGGGTTCTGTTAGGTACATTGCGCCTCCACCTCCTGGACCAGCACCGTATTTCATAAGCTTGCCTCCTCATGAAGATAAAATCTATCGATAA
Protein
MLLLLLLCLLPYIATQQQWVPCSELNDDLRYPCRCRVQVDRALQLRILMNCDRVVFAGDFPTLPYGAPIISFSQRWAGQQTLPMQIFSTYGLPLRELDFSHNSLRRLPDRLLAAVRGNLTKLALADNLLGDNLNPIFSTAELHNLPALEELDLSGNSIRGIEEGLLIGCDVLQVLRLDRNNINFIPSSSLNGPQSLRVLSLQENKIALLRQASFVSQKSLEEINLHGNMISTIEGGAFLGLKDLQILDLGRNRLSKFNSDVFQGAENLEKLDLSENFINDFPTVAIKTFGGLSHLNLSSNMISNIDNSHLNSLSSLQVLDLSRNNLVKLTPGTFVGLTELSYLDIGVNSLRTVEDNAFDGLTSLKTLLLRDNNILLIPATALSRLPNLVSVHLGFNRVTALSNDILRSVSDRVTSLVLSRNVIRELPTAAFDHFKMLRHLDLSGNLLNTIAAEVFDGLEQSLQFLSLSQNRILGFTGQQLKFVSLWFLDISGNQISNVPVEAFESIPSLTHLNMSRNQHINVLPQNLFQHNQALLTIDLSYNGLKALPVNLFSKNFNLQQIHLSNNLLQEISENTFKNLRNLTHLDLSYNNIVNLRTPSFVNVMSIQYLSLKGNQLSAFKGEFFNTGTSLEVIDLSDNQLSYLFPSAFKIHPRLREIKLANNKFNFFPSELISTLQYLELVDLSGNALKNVDELDFARLPKLRTILLARNELETVSEMAFHNSSQIQQLDLSQNKIDRLGDRLFEGLIRLETLNLAGNMLAELPDSIFERNRLHMLENINLSGNLFEHPPLKALQKQYFFVSAVDLSHNKIVDIPAEDSVLRLNISNNPIVMIMDGNFEGLTSLRVLNMNNLDKCTRIEKNAFRSLPNLIELRAFGYPRLGYFDVQGTLQYLFALEKLDVELKDTNIGPDQLHSTLHPRLEELGIRGSRLKTVSSGVLAGLKAPAIVVRLRNTSVSTLPPALLFPLPRSSQITIDVGGSQLETLQPQLLVALDDRRADLSMLGLDTNPIRCDCNARALRRWLPNVGIDNIRCQSPDYLSGYLLVEIGDDELTCDSKRRTTATSTSSVSTTSPPRLVHRTSSEPDIIWSVAPSHDRPKISGEPKGAPVIGVATASNDDNLIIGIVSGVVAFIAILIVAICIIRLKMTTTSYRGGPLANSPTGGAPAVWGPPWPGYTGTMPPASLSTATLPHKVQPGPGSVRYIAPPPPGPAPYFISLPPHEDKIYR
Summary
Uniprot
H9JAC2
A0A194R535
A0A194Q997
B2DBJ9
A0A0L7L0B1
A0A1Y1K8Z6
+ More
A0A1I8MB87 A0A2J7QE30 B4JS99 A0A0M4F7E4 B4KC35 A0A0L0C941 A0A1I8Q9R3 B4GNG1 Q29C89 A0A1B6EZ68 T1HRV9 J9K0G3 A0A3B0JJI1 A0A2H8TFD8 A0A0T6B441 A0A0L7RDL8 A0A0C9RAE7 A0A3L8DY72 A0A087ZZ13 A0A182G483 A0A182GHD0 A0A154PAT0 W5JU06 Q7QI68 A0A182VJ79 A0A182PQ82 A0A182ICZ6 A0A182WRX3 A0A182M525 A0A182KVT7 A0A182RQB6 A0A182IT29 A0A310SQA2 K7IYY6 E2B834 A0A182NQ89 E1ZUY0 A0A232ERP1 A0A182WK66 A0A182YP23 A0A084WJI3 A0A2P8YYJ0 A0A182K7Z6 A0A1I8N9L7 A0A182UFS5 A0A2M4AD16 A0A182S6D3 A0A1S4J269 A0A182Q478 A0A336MP89 A0A026VWW7 B0W204 B4HZI8 A0A2A3EDV1 A0A0K2TSS1 A0A182FD16 A0A147BJ45 A0A0N1ITC9 E0VD06
A0A1I8MB87 A0A2J7QE30 B4JS99 A0A0M4F7E4 B4KC35 A0A0L0C941 A0A1I8Q9R3 B4GNG1 Q29C89 A0A1B6EZ68 T1HRV9 J9K0G3 A0A3B0JJI1 A0A2H8TFD8 A0A0T6B441 A0A0L7RDL8 A0A0C9RAE7 A0A3L8DY72 A0A087ZZ13 A0A182G483 A0A182GHD0 A0A154PAT0 W5JU06 Q7QI68 A0A182VJ79 A0A182PQ82 A0A182ICZ6 A0A182WRX3 A0A182M525 A0A182KVT7 A0A182RQB6 A0A182IT29 A0A310SQA2 K7IYY6 E2B834 A0A182NQ89 E1ZUY0 A0A232ERP1 A0A182WK66 A0A182YP23 A0A084WJI3 A0A2P8YYJ0 A0A182K7Z6 A0A1I8N9L7 A0A182UFS5 A0A2M4AD16 A0A182S6D3 A0A1S4J269 A0A182Q478 A0A336MP89 A0A026VWW7 B0W204 B4HZI8 A0A2A3EDV1 A0A0K2TSS1 A0A182FD16 A0A147BJ45 A0A0N1ITC9 E0VD06
Pubmed
EMBL
BABH01009025
KQ460685
KPJ12918.1
KQ459551
KPI99995.1
AB264682
+ More
BAG30758.1 JTDY01003991 KOB68741.1 GEZM01088787 GEZM01088786 JAV57919.1 NEVH01015354 PNF26836.1 CH916373 EDV94639.1 CP012526 ALC47770.1 CH933806 EDW16908.2 JRES01000835 KNC27904.1 CH479186 EDW39287.1 CM000070 EAL26757.2 KRS99533.1 GECZ01026522 JAS43247.1 ACPB03010468 ABLF02014752 ABLF02017224 ABLF02017226 ABLF02017227 ABLF02017228 ABLF02017229 ABLF02037342 OUUW01000005 SPP80923.1 GFXV01001018 MBW12823.1 LJIG01009908 KRT82140.1 KQ414613 KOC68948.1 GBYB01005239 GBYB01005240 JAG75006.1 JAG75007.1 QOIP01000003 RLU25173.1 JXUM01041876 KQ561302 KXJ79061.1 JXUM01063401 KQ562247 KXJ76311.1 KQ434864 KZC09046.1 ADMH02000378 ETN66778.1 AAAB01008810 EAA04820.4 APCN01006079 AXCM01003177 KQ761768 OAD57105.1 GL446282 EFN88126.1 GL434326 EFN75015.1 NNAY01002583 OXU20991.1 ATLV01024035 KE525348 KFB50377.1 PYGN01000285 PSN49301.1 GGFK01005356 MBW38677.1 AXCN02001958 UFQT01001829 SSX31920.1 KK107652 EZA48288.1 DS231824 EDS27440.1 CH480819 EDW53445.1 KZ288269 PBC29925.1 HACA01011361 CDW28722.1 GEGO01004595 JAR90809.1 KQ435824 KOX72194.1 DS235070 EEB11262.1
BAG30758.1 JTDY01003991 KOB68741.1 GEZM01088787 GEZM01088786 JAV57919.1 NEVH01015354 PNF26836.1 CH916373 EDV94639.1 CP012526 ALC47770.1 CH933806 EDW16908.2 JRES01000835 KNC27904.1 CH479186 EDW39287.1 CM000070 EAL26757.2 KRS99533.1 GECZ01026522 JAS43247.1 ACPB03010468 ABLF02014752 ABLF02017224 ABLF02017226 ABLF02017227 ABLF02017228 ABLF02017229 ABLF02037342 OUUW01000005 SPP80923.1 GFXV01001018 MBW12823.1 LJIG01009908 KRT82140.1 KQ414613 KOC68948.1 GBYB01005239 GBYB01005240 JAG75006.1 JAG75007.1 QOIP01000003 RLU25173.1 JXUM01041876 KQ561302 KXJ79061.1 JXUM01063401 KQ562247 KXJ76311.1 KQ434864 KZC09046.1 ADMH02000378 ETN66778.1 AAAB01008810 EAA04820.4 APCN01006079 AXCM01003177 KQ761768 OAD57105.1 GL446282 EFN88126.1 GL434326 EFN75015.1 NNAY01002583 OXU20991.1 ATLV01024035 KE525348 KFB50377.1 PYGN01000285 PSN49301.1 GGFK01005356 MBW38677.1 AXCN02001958 UFQT01001829 SSX31920.1 KK107652 EZA48288.1 DS231824 EDS27440.1 CH480819 EDW53445.1 KZ288269 PBC29925.1 HACA01011361 CDW28722.1 GEGO01004595 JAR90809.1 KQ435824 KOX72194.1 DS235070 EEB11262.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000037510
UP000095301
UP000235965
+ More
UP000001070 UP000092553 UP000009192 UP000037069 UP000095300 UP000008744 UP000001819 UP000015103 UP000007819 UP000268350 UP000053825 UP000279307 UP000005203 UP000069940 UP000249989 UP000076502 UP000000673 UP000007062 UP000075903 UP000075885 UP000075840 UP000076407 UP000075883 UP000075882 UP000075900 UP000075880 UP000002358 UP000008237 UP000075884 UP000000311 UP000215335 UP000075920 UP000076408 UP000030765 UP000245037 UP000075881 UP000075902 UP000075901 UP000075886 UP000053097 UP000002320 UP000001292 UP000242457 UP000069272 UP000053105 UP000009046
UP000001070 UP000092553 UP000009192 UP000037069 UP000095300 UP000008744 UP000001819 UP000015103 UP000007819 UP000268350 UP000053825 UP000279307 UP000005203 UP000069940 UP000249989 UP000076502 UP000000673 UP000007062 UP000075903 UP000075885 UP000075840 UP000076407 UP000075883 UP000075882 UP000075900 UP000075880 UP000002358 UP000008237 UP000075884 UP000000311 UP000215335 UP000075920 UP000076408 UP000030765 UP000245037 UP000075881 UP000075902 UP000075901 UP000075886 UP000053097 UP000002320 UP000001292 UP000242457 UP000069272 UP000053105 UP000009046
PRIDE
Pfam
Interpro
IPR001611
Leu-rich_rpt
+ More
IPR003591 Leu-rich_rpt_typical-subtyp
IPR000483 Cys-rich_flank_reg_C
IPR032675 LRR_dom_sf
IPR026906 LRR_5
IPR025875 Leu-rich_rpt_4
IPR000626 Ubiquitin_dom
IPR029071 Ubiquitin-like_domsf
IPR022617 Rad60/SUMO-like_dom
IPR013083 Znf_RING/FYVE/PHD
IPR026850 FANCL_C
IPR019162 FancL_WD-rpt_cont_dom
IPR003591 Leu-rich_rpt_typical-subtyp
IPR000483 Cys-rich_flank_reg_C
IPR032675 LRR_dom_sf
IPR026906 LRR_5
IPR025875 Leu-rich_rpt_4
IPR000626 Ubiquitin_dom
IPR029071 Ubiquitin-like_domsf
IPR022617 Rad60/SUMO-like_dom
IPR013083 Znf_RING/FYVE/PHD
IPR026850 FANCL_C
IPR019162 FancL_WD-rpt_cont_dom
SUPFAM
SSF54236
SSF54236
Gene 3D
ProteinModelPortal
H9JAC2
A0A194R535
A0A194Q997
B2DBJ9
A0A0L7L0B1
A0A1Y1K8Z6
+ More
A0A1I8MB87 A0A2J7QE30 B4JS99 A0A0M4F7E4 B4KC35 A0A0L0C941 A0A1I8Q9R3 B4GNG1 Q29C89 A0A1B6EZ68 T1HRV9 J9K0G3 A0A3B0JJI1 A0A2H8TFD8 A0A0T6B441 A0A0L7RDL8 A0A0C9RAE7 A0A3L8DY72 A0A087ZZ13 A0A182G483 A0A182GHD0 A0A154PAT0 W5JU06 Q7QI68 A0A182VJ79 A0A182PQ82 A0A182ICZ6 A0A182WRX3 A0A182M525 A0A182KVT7 A0A182RQB6 A0A182IT29 A0A310SQA2 K7IYY6 E2B834 A0A182NQ89 E1ZUY0 A0A232ERP1 A0A182WK66 A0A182YP23 A0A084WJI3 A0A2P8YYJ0 A0A182K7Z6 A0A1I8N9L7 A0A182UFS5 A0A2M4AD16 A0A182S6D3 A0A1S4J269 A0A182Q478 A0A336MP89 A0A026VWW7 B0W204 B4HZI8 A0A2A3EDV1 A0A0K2TSS1 A0A182FD16 A0A147BJ45 A0A0N1ITC9 E0VD06
A0A1I8MB87 A0A2J7QE30 B4JS99 A0A0M4F7E4 B4KC35 A0A0L0C941 A0A1I8Q9R3 B4GNG1 Q29C89 A0A1B6EZ68 T1HRV9 J9K0G3 A0A3B0JJI1 A0A2H8TFD8 A0A0T6B441 A0A0L7RDL8 A0A0C9RAE7 A0A3L8DY72 A0A087ZZ13 A0A182G483 A0A182GHD0 A0A154PAT0 W5JU06 Q7QI68 A0A182VJ79 A0A182PQ82 A0A182ICZ6 A0A182WRX3 A0A182M525 A0A182KVT7 A0A182RQB6 A0A182IT29 A0A310SQA2 K7IYY6 E2B834 A0A182NQ89 E1ZUY0 A0A232ERP1 A0A182WK66 A0A182YP23 A0A084WJI3 A0A2P8YYJ0 A0A182K7Z6 A0A1I8N9L7 A0A182UFS5 A0A2M4AD16 A0A182S6D3 A0A1S4J269 A0A182Q478 A0A336MP89 A0A026VWW7 B0W204 B4HZI8 A0A2A3EDV1 A0A0K2TSS1 A0A182FD16 A0A147BJ45 A0A0N1ITC9 E0VD06
PDB
4KT1
E-value=7.96586e-32,
Score=347
Ontologies
GO
Topology
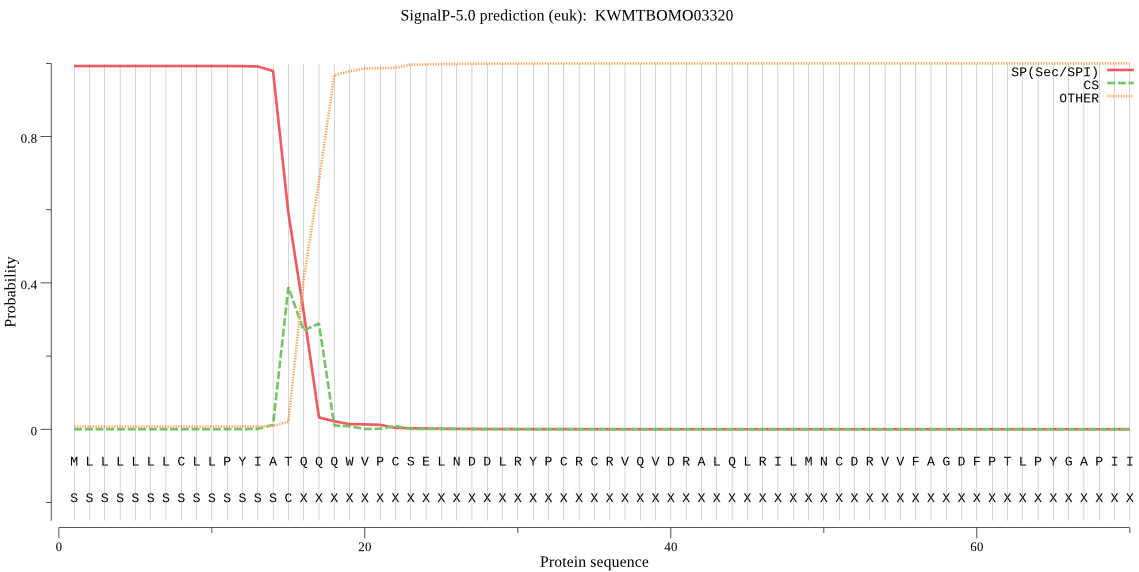
SignalP
Position: 1 - 15,
Likelihood: 0.992517
Length:
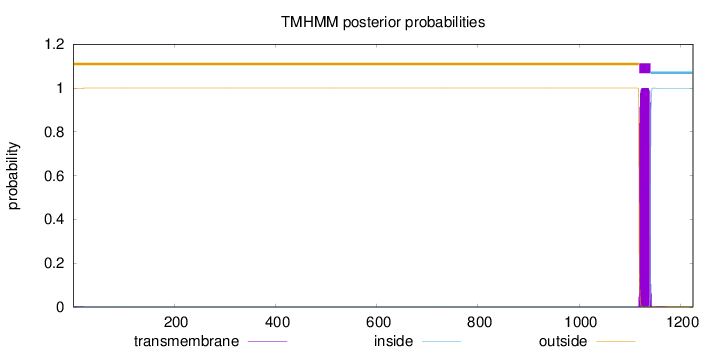
1225
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.93356
Exp number, first 60 AAs:
0.03491
Total prob of N-in:
0.00191
outside
1 - 1118
TMhelix
1119 - 1141
inside
1142 - 1225
Population Genetic Test Statistics
Pi
166.949695
Theta
163.871798
Tajima's D
0.070996
CLR
1.277931
CSRT
0.394480275986201
Interpretation
Uncertain
