Pre Gene Modal
BGIBMGA006411
Annotation
PREDICTED:_vacuolar_protein_sorting-associated_protein_16_homolog_[Plutella_xylostella]
Full name
Vacuolar protein sorting-associated protein 16 homolog
Location in the cell
Cytoplasmic Reliability : 2.098
Sequence
CDS
ATGTCGGCTCTACTAACAGCTGATTGGTTCCAACTGGACACATATTATCGAAAATTTGATCTCTACACAATGAACTGGATGATGGACGAAGGTTTAGAGAATATGATAGTTAGCGGTGCACCGTATGGTGGCCCAATGGCAGTAGTGAGAGACCGTAAACAATTTGTTCAAATAAAAACCACCACTAAACCTGTTATAACAATCTACAACTGCTCAGGAAATGTTATATCAAAGATATTGTGGAACAGTGGTGTCCTACTACATATCGGATGGTCTGATGGGGAACAGCTACTTTGTATTCAAGAAAGCGGTGATGTCCTTATTTATGACATGTTTGGAGCTTACCAGAAAACTTTCAATATGGGCCAAGAAGTCAGAGATACAAAAGTTTGTAAAGCTCAATTGTTTCCAAATCCTCATGGTATAGGACTAGCTGTTATCACTACAACTAATAGAATGTTCCTTGTGAGTAATGTTTCTGAACCAAAAGCTCGACCGGTTCCTGATATACCAAGAGCCAATGAACCGATATCATGCTGGTGCGCTGTAAACTCTAGTTTTATAGTGTGTCGAGATAAAGAGATTTATAAATGCCAACTAGGTGAATCCAGGGCCATTCTTTTGCGTCCAGAAATCAAGAACCCCTACACACAGATCCTATCTATCGTGGCTTCACAGAATGGCAAGCACATTGCGTTCTTTACCGACTCTGGTTTCCTATGGATTGGATCTTCCGATTTGAGGAATAATTACTGCGAGTTAGATACGGATTATATTAAACAACCCAAAGAATTCATGTGGTGCGGTTCCCAAGCTGTGGCCGCACACTGGGACGACACTTTGTGCATGTACGGTACTAAAGGCAATAGTGTTGCGTATCCATACGATGGACCCTTCCACTTAATTCAGGAGATGGACTGCATACGTATTGTATCAGAAATGACCCATGAACTCGTGCAAAAGGTGCCTTTTGTTGTTGAGAAAATATTCCGGATCAATAGCACGGCGCCCGGCTCGTACCTAGTTGAAGCTTCTAAGCAATTTCAGAAACGCAGTCACCGAGCTGACGAGTACATCCGGCTGGTGAAGCCCGACCTGTCCGCCGCCGTCGCGGACTGCATCGAGGCAGCCGCCTTCGAGTTCAGTCCGGACGTGCAGAAAATGTTGATCCGGGCCGCGCAGTTCGGGAAATGCTTCCTGATGGACCCCGTCATCACGGAGCTCTACGTCAAGACGTGTCGCTGGTTGAGAGTTCTGAACGCCGTCAGAGATCCTAAAGTGGCTATACCTTTGACGTCTATGCAAATGCGTAATTTGGGCGAACGAATACTTCTTGACCGGCTCATATGGCGCCGACTGCACTGTTTGGCCGGACACATCGCTTCCTACCTGCAACTGAAGGATGGACGCACGAGAGTCCTGTCCCATTGGGCTTGTTACAAGGTAACTCAGCCTCACTTGGACAACGAGAGTGCCGCAAGGGAGATAGGCGAGAAGCTACGGAATGTCCCTGGTATATCGTACACAACGATAGCCATGAAAGCTGCCGAAAAAGGAAGGAAAGCTCTCGCTATAAAAATCCTGGAGTACGAGGCACATAGCAAATTACAAGTTCCATTACTATTGTCACTCGGCGAAGGTCCGACTGCGCTGCTGAAGGCGACCGCCAGCGGCGACACGGACTTGGTCTACGTTGTTCTGTTGCATCTCAAGGAAAAAATGGGAAAACATGACTTTGAGCTCACGATCCGCAGTTTCCCTTTAGCGCACGCCCTGTACATCAAGTACTGCGCAAGTCACAACCGCGAGGCCCTGCGCCTGGTCTACGTTCAGGAGGACGACTTCCAGGGTCAGGCCGCCACTCACATATGTGACGCCATCGAGCAGACCAACCCGGGCAGCGCGGAGGCCTCGCTGATATCCGCCAGGGAATGTTACAAAAAGGGCAAAAACGAATTAGGTGCATCTGTATGCGAGGACGCCCGGAAGCTGTGCAAGCAACAGTCCAGTCTCCAGGAGACGTACGGAACGAGCTTCGTGGGGCTGTCGCTTCACGATACCGTCAAAAAGCTTCTGGACCAAGGAGAAGTCAGATTGGCGGACAAACTGCGATCGGAATATAAAATGCCCGATCGAAGATATTGGTGGCTTCGTATCCTAACACTAGCCGAGAAGTCCAATTGGGAGGAATTGGATAAGTTTTCGAAATCAAAGAAATCTCCAGTCGGGTACGAGCCGTTTGTCGACGCCTGTCTCAAGTACAACAAGGCGGACGAAGCACTCAAGTACTTGCCGAGATGCAGGGATGAGATCAAAATTAAATACTACGTCAAAGCTGGGTATTACGCGGAGGCCGCCGAAGTAGCATACGAACAGAAAGACGAAGCCGGCTTACATTTCGTTCAAAACAAATGCCCGATGCAAGACTTCGACAATCAAGCCAAAATATCAGGACTCCTAGAACAGTTGAAAATTAAGAAATGA
Protein
MSALLTADWFQLDTYYRKFDLYTMNWMMDEGLENMIVSGAPYGGPMAVVRDRKQFVQIKTTTKPVITIYNCSGNVISKILWNSGVLLHIGWSDGEQLLCIQESGDVLIYDMFGAYQKTFNMGQEVRDTKVCKAQLFPNPHGIGLAVITTTNRMFLVSNVSEPKARPVPDIPRANEPISCWCAVNSSFIVCRDKEIYKCQLGESRAILLRPEIKNPYTQILSIVASQNGKHIAFFTDSGFLWIGSSDLRNNYCELDTDYIKQPKEFMWCGSQAVAAHWDDTLCMYGTKGNSVAYPYDGPFHLIQEMDCIRIVSEMTHELVQKVPFVVEKIFRINSTAPGSYLVEASKQFQKRSHRADEYIRLVKPDLSAAVADCIEAAAFEFSPDVQKMLIRAAQFGKCFLMDPVITELYVKTCRWLRVLNAVRDPKVAIPLTSMQMRNLGERILLDRLIWRRLHCLAGHIASYLQLKDGRTRVLSHWACYKVTQPHLDNESAAREIGEKLRNVPGISYTTIAMKAAEKGRKALAIKILEYEAHSKLQVPLLLSLGEGPTALLKATASGDTDLVYVVLLHLKEKMGKHDFELTIRSFPLAHALYIKYCASHNREALRLVYVQEDDFQGQAATHICDAIEQTNPGSAEASLISARECYKKGKNELGASVCEDARKLCKQQSSLQETYGTSFVGLSLHDTVKKLLDQGEVRLADKLRSEYKMPDRRYWWLRILTLAEKSNWEELDKFSKSKKSPVGYEPFVDACLKYNKADEALKYLPRCRDEIKIKYYVKAGYYAEAAEVAYEQKDEAGLHFVQNKCPMQDFDNQAKISGLLEQLKIKK
Summary
Description
Plays a role in vesicle-mediated protein trafficking to lysosomal compartments including the endocytic membrane transport and autophagic pathways. Believed to act as a core component of the putative HOPS and CORVET endosomal tethering complexes.
Similarity
Belongs to the VPS16 family.
Uniprot
H9JA66
A0A194R5G4
A0A194Q381
A0A2A4ITH3
A0A1E1WMG7
A0A067RW40
+ More
A0A2J7RED3 D7GXI5 A0A1W4XG90 F4WMZ2 A0A195FUF2 A0A158NSJ7 A0A151WIB8 A0A151HYW3 E2A495 A0A0C9QK40 E9JD91 A0A1Y1NG99 A0A151J431 E2B833 A0A0L7RDQ4 A0A1B6KXY6 A0A232F043 A0A310SG39 A0A154PAW8 A0A0C9R7F3 A0A2A3EFJ4 A0A023F2B4 A0A087ZYS2 A0A069DX66 E0VS77 A0A0P4VTH1 T1HR96 A0A1B0GLC8 R7VKD8 A0A1S3I6Y6 A0A336MYR0 A0A1L8DVT0 A0A1D2MTX4 A0A0P6FJG8 A0A0P5PBJ2 A0A087TTA1 A0A164Y9W7 A0A0P6GMG9 A0A0P5U117 A0A0P5B543 E9HJU2 A0A0P6EPB5 A0A226D3J7 A0A0A9X2X7 W4ZC77 A0A0P5RPQ7 A0A0P4YGN6 A0A0B7AXF7 A0A0P6I948 A0A0N8DNG6 A0A1J1I0H2 A0A0P5B285 B0W200 A0A1Q3FAJ4 K1PWU7 Q16SL9 A0A131YNZ8 A0A0N8AT84 A0A224ZBT6 A0A146LUE5 A0A182G9E2 A0A131XNR4 A0A0P5DTV2 A0A0P5N1E1 A0A210PVW1 A0A1A9WZ26 A0A0L0CIS0 A0A0P5CWU8 V4AP47 A0A1I8Q4M5 A0A0P5U1Z6 A0A0P6G4M8 W8BDD4 A0A182YMX9 A0A2S2QA28 A0A2B4S3T7 A0A182R9J2 A0A1I8NI14 A0A182ILF1 A0A182K7D2 A0A0P6ES34 A0A0A1XM34 J9JWL8 A0A1E1XSW3 A0A2T7PR81 A0A1B0BHP4 A0A084VIW0 A0A182UXI3 A0A182SFP9 A0A131Y4C9 A0A182KYB5 Q7QFJ5 B4K623
A0A2J7RED3 D7GXI5 A0A1W4XG90 F4WMZ2 A0A195FUF2 A0A158NSJ7 A0A151WIB8 A0A151HYW3 E2A495 A0A0C9QK40 E9JD91 A0A1Y1NG99 A0A151J431 E2B833 A0A0L7RDQ4 A0A1B6KXY6 A0A232F043 A0A310SG39 A0A154PAW8 A0A0C9R7F3 A0A2A3EFJ4 A0A023F2B4 A0A087ZYS2 A0A069DX66 E0VS77 A0A0P4VTH1 T1HR96 A0A1B0GLC8 R7VKD8 A0A1S3I6Y6 A0A336MYR0 A0A1L8DVT0 A0A1D2MTX4 A0A0P6FJG8 A0A0P5PBJ2 A0A087TTA1 A0A164Y9W7 A0A0P6GMG9 A0A0P5U117 A0A0P5B543 E9HJU2 A0A0P6EPB5 A0A226D3J7 A0A0A9X2X7 W4ZC77 A0A0P5RPQ7 A0A0P4YGN6 A0A0B7AXF7 A0A0P6I948 A0A0N8DNG6 A0A1J1I0H2 A0A0P5B285 B0W200 A0A1Q3FAJ4 K1PWU7 Q16SL9 A0A131YNZ8 A0A0N8AT84 A0A224ZBT6 A0A146LUE5 A0A182G9E2 A0A131XNR4 A0A0P5DTV2 A0A0P5N1E1 A0A210PVW1 A0A1A9WZ26 A0A0L0CIS0 A0A0P5CWU8 V4AP47 A0A1I8Q4M5 A0A0P5U1Z6 A0A0P6G4M8 W8BDD4 A0A182YMX9 A0A2S2QA28 A0A2B4S3T7 A0A182R9J2 A0A1I8NI14 A0A182ILF1 A0A182K7D2 A0A0P6ES34 A0A0A1XM34 J9JWL8 A0A1E1XSW3 A0A2T7PR81 A0A1B0BHP4 A0A084VIW0 A0A182UXI3 A0A182SFP9 A0A131Y4C9 A0A182KYB5 Q7QFJ5 B4K623
Pubmed
19121390
26354079
24845553
18362917
19820115
21719571
+ More
21347285 20798317 21282665 28004739 28648823 25474469 26334808 20566863 27129103 23254933 27289101 21292972 25401762 22992520 17510324 26830274 28797301 26823975 26483478 28049606 28812685 26108605 24495485 25244985 25315136 25830018 29209593 24438588 20966253 12364791 14747013 17210077 17994087
21347285 20798317 21282665 28004739 28648823 25474469 26334808 20566863 27129103 23254933 27289101 21292972 25401762 22992520 17510324 26830274 28797301 26823975 26483478 28049606 28812685 26108605 24495485 25244985 25315136 25830018 29209593 24438588 20966253 12364791 14747013 17210077 17994087
EMBL
BABH01009017
BABH01009018
KQ460685
KPJ12917.1
KQ459551
KPI99996.1
+ More
NWSH01008438 PCG62554.1 GDQN01002841 JAT88213.1 KK852427 KDR24089.1 NEVH01005277 PNF39187.1 KQ971382 EFA13302.2 GL888223 EGI64462.1 KQ981261 KYN44083.1 ADTU01024980 KQ983089 KYQ47592.1 KQ976720 KYM76718.1 GL436596 EFN71766.1 GBYB01003919 JAG73686.1 GL771866 EFZ09129.1 GEZM01003788 GEZM01003787 GEZM01003786 GEZM01003785 JAV96568.1 KQ980211 KYN17190.1 GL446282 EFN88125.1 KQ414613 KOC68949.1 GEBQ01023670 JAT16307.1 NNAY01001413 OXU24054.1 KQ761768 OAD57104.1 KQ434864 KZC09045.1 GBYB01003920 JAG73687.1 KZ288269 PBC29926.1 GBBI01003583 JAC15129.1 GBGD01000488 JAC88401.1 DS235745 EEB16233.1 GDKW01000769 JAI55826.1 ACPB03014376 AJWK01034746 AMQN01016953 KB292583 ELU17286.1 UFQS01003893 UFQT01003893 SSX16017.1 SSX35350.1 GFDF01003546 JAV10538.1 LJIJ01000528 ODM96537.1 GDIQ01063445 JAN31292.1 GDIQ01152610 GDIQ01136571 GDIQ01073931 JAK99115.1 KK116633 KFM68340.1 LRGB01000930 KZS15033.1 GDIQ01031470 JAN63267.1 GDIP01119281 JAL84433.1 GDIP01189516 JAJ33886.1 GL732664 EFX68012.1 GDIQ01059745 JAN34992.1 LNIX01000040 OXA39231.1 GBHO01029250 JAG14354.1 AAGJ04016044 AAGJ04016045 GDIQ01106623 JAL45103.1 GDIP01227915 JAI95486.1 HACG01037725 CEK84590.1 GDIQ01009513 JAN85224.1 GDIP01016519 JAM87196.1 CVRI01000036 CRK93076.1 GDIP01195167 JAJ28235.1 DS231824 EDS27436.1 GFDL01010441 JAV24604.1 JH816093 EKC23509.1 CH477672 EAT37446.1 GEDV01007870 JAP80687.1 GDIQ01245187 JAK06538.1 GFPF01013224 MAA24370.1 GDHC01008277 JAQ10352.1 JXUM01050240 KQ561640 KXJ77917.1 GEFH01000344 JAP68237.1 GDIP01151670 JAJ71732.1 GDIQ01148472 JAL03254.1 NEDP02005459 OWF40606.1 JRES01000338 KNC32150.1 GDIP01164534 JAJ58868.1 KB201459 ESO96565.1 GDIP01119282 JAL84432.1 GDIQ01057145 JAN37592.1 GAMC01009853 JAB96702.1 GGMS01005395 MBY74598.1 LSMT01000173 PFX24561.1 GDIQ01059744 JAN34993.1 GBXI01007435 GBXI01002226 JAD06857.1 JAD12066.1 ABLF02018142 ABLF02018146 GFAA01001036 JAU02399.1 PZQS01000002 PVD35929.1 JXJN01014484 ATLV01013411 KE524855 KFB37904.1 GEFM01002890 JAP72906.1 AAAB01008846 EAA06254.3 CH933806 EDW16260.1
NWSH01008438 PCG62554.1 GDQN01002841 JAT88213.1 KK852427 KDR24089.1 NEVH01005277 PNF39187.1 KQ971382 EFA13302.2 GL888223 EGI64462.1 KQ981261 KYN44083.1 ADTU01024980 KQ983089 KYQ47592.1 KQ976720 KYM76718.1 GL436596 EFN71766.1 GBYB01003919 JAG73686.1 GL771866 EFZ09129.1 GEZM01003788 GEZM01003787 GEZM01003786 GEZM01003785 JAV96568.1 KQ980211 KYN17190.1 GL446282 EFN88125.1 KQ414613 KOC68949.1 GEBQ01023670 JAT16307.1 NNAY01001413 OXU24054.1 KQ761768 OAD57104.1 KQ434864 KZC09045.1 GBYB01003920 JAG73687.1 KZ288269 PBC29926.1 GBBI01003583 JAC15129.1 GBGD01000488 JAC88401.1 DS235745 EEB16233.1 GDKW01000769 JAI55826.1 ACPB03014376 AJWK01034746 AMQN01016953 KB292583 ELU17286.1 UFQS01003893 UFQT01003893 SSX16017.1 SSX35350.1 GFDF01003546 JAV10538.1 LJIJ01000528 ODM96537.1 GDIQ01063445 JAN31292.1 GDIQ01152610 GDIQ01136571 GDIQ01073931 JAK99115.1 KK116633 KFM68340.1 LRGB01000930 KZS15033.1 GDIQ01031470 JAN63267.1 GDIP01119281 JAL84433.1 GDIP01189516 JAJ33886.1 GL732664 EFX68012.1 GDIQ01059745 JAN34992.1 LNIX01000040 OXA39231.1 GBHO01029250 JAG14354.1 AAGJ04016044 AAGJ04016045 GDIQ01106623 JAL45103.1 GDIP01227915 JAI95486.1 HACG01037725 CEK84590.1 GDIQ01009513 JAN85224.1 GDIP01016519 JAM87196.1 CVRI01000036 CRK93076.1 GDIP01195167 JAJ28235.1 DS231824 EDS27436.1 GFDL01010441 JAV24604.1 JH816093 EKC23509.1 CH477672 EAT37446.1 GEDV01007870 JAP80687.1 GDIQ01245187 JAK06538.1 GFPF01013224 MAA24370.1 GDHC01008277 JAQ10352.1 JXUM01050240 KQ561640 KXJ77917.1 GEFH01000344 JAP68237.1 GDIP01151670 JAJ71732.1 GDIQ01148472 JAL03254.1 NEDP02005459 OWF40606.1 JRES01000338 KNC32150.1 GDIP01164534 JAJ58868.1 KB201459 ESO96565.1 GDIP01119282 JAL84432.1 GDIQ01057145 JAN37592.1 GAMC01009853 JAB96702.1 GGMS01005395 MBY74598.1 LSMT01000173 PFX24561.1 GDIQ01059744 JAN34993.1 GBXI01007435 GBXI01002226 JAD06857.1 JAD12066.1 ABLF02018142 ABLF02018146 GFAA01001036 JAU02399.1 PZQS01000002 PVD35929.1 JXJN01014484 ATLV01013411 KE524855 KFB37904.1 GEFM01002890 JAP72906.1 AAAB01008846 EAA06254.3 CH933806 EDW16260.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000027135
UP000235965
+ More
UP000007266 UP000192223 UP000007755 UP000078541 UP000005205 UP000075809 UP000078540 UP000000311 UP000078492 UP000008237 UP000053825 UP000215335 UP000076502 UP000242457 UP000005203 UP000009046 UP000015103 UP000092461 UP000014760 UP000085678 UP000094527 UP000054359 UP000076858 UP000000305 UP000198287 UP000007110 UP000183832 UP000002320 UP000005408 UP000008820 UP000069940 UP000249989 UP000242188 UP000091820 UP000037069 UP000030746 UP000095300 UP000076408 UP000225706 UP000075900 UP000095301 UP000075880 UP000075881 UP000007819 UP000245119 UP000092460 UP000030765 UP000075903 UP000075901 UP000075882 UP000007062 UP000009192
UP000007266 UP000192223 UP000007755 UP000078541 UP000005205 UP000075809 UP000078540 UP000000311 UP000078492 UP000008237 UP000053825 UP000215335 UP000076502 UP000242457 UP000005203 UP000009046 UP000015103 UP000092461 UP000014760 UP000085678 UP000094527 UP000054359 UP000076858 UP000000305 UP000198287 UP000007110 UP000183832 UP000002320 UP000005408 UP000008820 UP000069940 UP000249989 UP000242188 UP000091820 UP000037069 UP000030746 UP000095300 UP000076408 UP000225706 UP000075900 UP000095301 UP000075880 UP000075881 UP000007819 UP000245119 UP000092460 UP000030765 UP000075903 UP000075901 UP000075882 UP000007062 UP000009192
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JA66
A0A194R5G4
A0A194Q381
A0A2A4ITH3
A0A1E1WMG7
A0A067RW40
+ More
A0A2J7RED3 D7GXI5 A0A1W4XG90 F4WMZ2 A0A195FUF2 A0A158NSJ7 A0A151WIB8 A0A151HYW3 E2A495 A0A0C9QK40 E9JD91 A0A1Y1NG99 A0A151J431 E2B833 A0A0L7RDQ4 A0A1B6KXY6 A0A232F043 A0A310SG39 A0A154PAW8 A0A0C9R7F3 A0A2A3EFJ4 A0A023F2B4 A0A087ZYS2 A0A069DX66 E0VS77 A0A0P4VTH1 T1HR96 A0A1B0GLC8 R7VKD8 A0A1S3I6Y6 A0A336MYR0 A0A1L8DVT0 A0A1D2MTX4 A0A0P6FJG8 A0A0P5PBJ2 A0A087TTA1 A0A164Y9W7 A0A0P6GMG9 A0A0P5U117 A0A0P5B543 E9HJU2 A0A0P6EPB5 A0A226D3J7 A0A0A9X2X7 W4ZC77 A0A0P5RPQ7 A0A0P4YGN6 A0A0B7AXF7 A0A0P6I948 A0A0N8DNG6 A0A1J1I0H2 A0A0P5B285 B0W200 A0A1Q3FAJ4 K1PWU7 Q16SL9 A0A131YNZ8 A0A0N8AT84 A0A224ZBT6 A0A146LUE5 A0A182G9E2 A0A131XNR4 A0A0P5DTV2 A0A0P5N1E1 A0A210PVW1 A0A1A9WZ26 A0A0L0CIS0 A0A0P5CWU8 V4AP47 A0A1I8Q4M5 A0A0P5U1Z6 A0A0P6G4M8 W8BDD4 A0A182YMX9 A0A2S2QA28 A0A2B4S3T7 A0A182R9J2 A0A1I8NI14 A0A182ILF1 A0A182K7D2 A0A0P6ES34 A0A0A1XM34 J9JWL8 A0A1E1XSW3 A0A2T7PR81 A0A1B0BHP4 A0A084VIW0 A0A182UXI3 A0A182SFP9 A0A131Y4C9 A0A182KYB5 Q7QFJ5 B4K623
A0A2J7RED3 D7GXI5 A0A1W4XG90 F4WMZ2 A0A195FUF2 A0A158NSJ7 A0A151WIB8 A0A151HYW3 E2A495 A0A0C9QK40 E9JD91 A0A1Y1NG99 A0A151J431 E2B833 A0A0L7RDQ4 A0A1B6KXY6 A0A232F043 A0A310SG39 A0A154PAW8 A0A0C9R7F3 A0A2A3EFJ4 A0A023F2B4 A0A087ZYS2 A0A069DX66 E0VS77 A0A0P4VTH1 T1HR96 A0A1B0GLC8 R7VKD8 A0A1S3I6Y6 A0A336MYR0 A0A1L8DVT0 A0A1D2MTX4 A0A0P6FJG8 A0A0P5PBJ2 A0A087TTA1 A0A164Y9W7 A0A0P6GMG9 A0A0P5U117 A0A0P5B543 E9HJU2 A0A0P6EPB5 A0A226D3J7 A0A0A9X2X7 W4ZC77 A0A0P5RPQ7 A0A0P4YGN6 A0A0B7AXF7 A0A0P6I948 A0A0N8DNG6 A0A1J1I0H2 A0A0P5B285 B0W200 A0A1Q3FAJ4 K1PWU7 Q16SL9 A0A131YNZ8 A0A0N8AT84 A0A224ZBT6 A0A146LUE5 A0A182G9E2 A0A131XNR4 A0A0P5DTV2 A0A0P5N1E1 A0A210PVW1 A0A1A9WZ26 A0A0L0CIS0 A0A0P5CWU8 V4AP47 A0A1I8Q4M5 A0A0P5U1Z6 A0A0P6G4M8 W8BDD4 A0A182YMX9 A0A2S2QA28 A0A2B4S3T7 A0A182R9J2 A0A1I8NI14 A0A182ILF1 A0A182K7D2 A0A0P6ES34 A0A0A1XM34 J9JWL8 A0A1E1XSW3 A0A2T7PR81 A0A1B0BHP4 A0A084VIW0 A0A182UXI3 A0A182SFP9 A0A131Y4C9 A0A182KYB5 Q7QFJ5 B4K623
PDB
5BV1
E-value=5.4052e-37,
Score=390
Ontologies
GO
PANTHER
Topology
Subcellular location
Late endosome membrane
Cytoplasmic, peripheral membrane protein associated with late endosomes/lysosomes. With evidence from 1 publications.
Lysosome membrane Cytoplasmic, peripheral membrane protein associated with late endosomes/lysosomes. With evidence from 1 publications.
Lysosome membrane Cytoplasmic, peripheral membrane protein associated with late endosomes/lysosomes. With evidence from 1 publications.
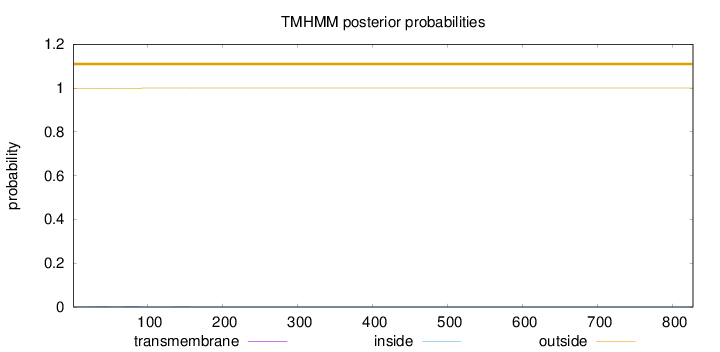
Length:
827
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0700000000000001
Exp number, first 60 AAs:
0.0298
Total prob of N-in:
0.00188
outside
1 - 827
Population Genetic Test Statistics
Pi
195.396331
Theta
168.296407
Tajima's D
0.527904
CLR
0
CSRT
0.517174141292935
Interpretation
Uncertain
