Gene
KWMTBOMO03318
Pre Gene Modal
BGIBMGA006466
Annotation
PREDICTED:_juxtaposed_with_another_zinc_finger_protein_1_[Amyelois_transitella]
Transcription factor
Location in the cell
Nuclear Reliability : 3.937
Sequence
CDS
ATGGCCGTCTTCATGATTAACATCTGTAAATTTAATGACTGTGGAATTATATTTCCAAGATTAACACAACTTATTGAACACATCGAGGATATTCATATAGATTACGATCCGGAGGTGGTAGAGCAGAAAGAGGCTTCCCAGCCGTCATGCATCCCACTAAGTTACATTTTGAAATATATATCAGAAGCGTCAAGACGCGAAAATCACAATGCACCTCCTGTTCCGGAGGTCCGCCGGCGTTTACTCTCCGCACCCAAAACACCATCAGTGCGCAGCAGTACACCCACAGGAAGTGAAATGGATGAAGACGAGGCAGGTACTCATTCAGAGGACAGTAATGACTCATGGACAACGGTAGAAGAGTACAGCTCTGAGTTCATATTGCGTTACGGGATGAGAATGAATCCAAACGCCTTGTCCGGGGCTTTGGGTCCTGTCGCTCAAGAGAAGCCATTTGCTTGCCCAGTACCTGGCTGCAAGAAGCGATACAAGAACGTCAACGGCATCAAGTACCACTCTAAGAACGGTCACAAGAAGGACGGCAAGGTAAGAAAGGCTTACAAATGCCAGTGCGGCAAAAGTTACAAAACCGCCCAAGGATTTAAGAGCCACTCCATATCGCAACACATCGCCAATAGCACAGTGCTGGCGGTCCCATCAGAGCTGGTCCGGCCGGCGCAGAGAATGCCCGCCGTCGTGGTGACGGCGTCGGGCTCTCGTCCGCACCACGGCCCCGACGGAGCTGACAGCAACAAGCTGGTTCGCATCTATGACACACTAAAAAACAAAGATCTCTCATCGTTTACCATCACTAAAAGTAATCTCGGCATCAATGTCCTATCGAAGCAAAGTCATTTACGTCACGGTCTCCCGTCGTCTCACTCCCCGCCCGTTACGAGTCTCGTTGAGAAAAATGTCGACGCTTATACTAAAGTGACGGTGCAGAGCGCCCAACAGGTTTACGCACAGTCTTTCACTAATGAGACGGCCAATTAA
Protein
MAVFMINICKFNDCGIIFPRLTQLIEHIEDIHIDYDPEVVEQKEASQPSCIPLSYILKYISEASRRENHNAPPVPEVRRRLLSAPKTPSVRSSTPTGSEMDEDEAGTHSEDSNDSWTTVEEYSSEFILRYGMRMNPNALSGALGPVAQEKPFACPVPGCKKRYKNVNGIKYHSKNGHKKDGKVRKAYKCQCGKSYKTAQGFKSHSISQHIANSTVLAVPSELVRPAQRMPAVVVTASGSRPHHGPDGADSNKLVRIYDTLKNKDLSSFTITKSNLGINVLSKQSHLRHGLPSSHSPPVTSLVEKNVDAYTKVTVQSAQQVYAQSFTNETAN
Summary
Uniprot
H9JAC1
A0A194RAM9
A0A2A4J3T1
A0A194Q365
A0A2H1W6R9
A0A212FKY3
+ More
A0A0K8UZ63 A0A0K8WE69 A0A034V7V7 A0A0K8VEP6 A0A2P8XF54 B4PNB5 Q9V9Z6 A0A0B4K810 A0A0R1EBU8 A0A0A1XBI3 A0A2J7REB6 B4GN56 Q29CE9 A0A1W4UI25 A0A1W4UHZ4 B3M1K9 A0A0P9CA52 I5AMT0 B3P7R6 A0A3B0JKG7 B4I004 A0A0M4EHF6 B4QSK6 A0A3B0JEK7 A0A067RM36 E0VS75 B4NFC6 A0A0Q9X1F7 A0A232EU34 K7J2T0 B4K625 B4M5U6 A0A1B6BW75 B4JYP0 A0A1A9Z2E6 A0A224Z0D4 A0A1E1XGN9 A0A131YW98 L7LXQ2 A0A023G7G5 A0A3P8V531 A0A2U9AYQ9 A0A060XM74 A0A1S3M607 A0A3B4Y6A1 A0A3B4T5T0 A0A1S3Q1J8 H3DM52 Q1KKZ9 A0A147AAM3 A0A3P8NKD2 A0A3P9C9G1 A0A3B4GDS5 A8DSV7 I3J4A5 A0A3B5N0D4 A0A087XEK7
A0A0K8UZ63 A0A0K8WE69 A0A034V7V7 A0A0K8VEP6 A0A2P8XF54 B4PNB5 Q9V9Z6 A0A0B4K810 A0A0R1EBU8 A0A0A1XBI3 A0A2J7REB6 B4GN56 Q29CE9 A0A1W4UI25 A0A1W4UHZ4 B3M1K9 A0A0P9CA52 I5AMT0 B3P7R6 A0A3B0JKG7 B4I004 A0A0M4EHF6 B4QSK6 A0A3B0JEK7 A0A067RM36 E0VS75 B4NFC6 A0A0Q9X1F7 A0A232EU34 K7J2T0 B4K625 B4M5U6 A0A1B6BW75 B4JYP0 A0A1A9Z2E6 A0A224Z0D4 A0A1E1XGN9 A0A131YW98 L7LXQ2 A0A023G7G5 A0A3P8V531 A0A2U9AYQ9 A0A060XM74 A0A1S3M607 A0A3B4Y6A1 A0A3B4T5T0 A0A1S3Q1J8 H3DM52 Q1KKZ9 A0A147AAM3 A0A3P8NKD2 A0A3P9C9G1 A0A3B4GDS5 A8DSV7 I3J4A5 A0A3B5N0D4 A0A087XEK7
Pubmed
19121390
26354079
22118469
25348373
29403074
17994087
+ More
17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 15632085 24845553 20566863 28648823 20075255 28797301 28503490 26830274 25576852 24487278 24755649 15496914 16636282 21551351 25186727 17845724
17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 15632085 24845553 20566863 28648823 20075255 28797301 28503490 26830274 25576852 24487278 24755649 15496914 16636282 21551351 25186727 17845724
EMBL
BABH01009016
KQ460685
KPJ12916.1
NWSH01003198
PCG66807.1
KQ459551
+ More
KPI99997.1 ODYU01006696 SOQ48770.1 AGBW02007989 OWR54349.1 GDHF01020486 JAI31828.1 GDHF01002932 JAI49382.1 GAKP01019566 JAC39386.1 GDHF01015022 JAI37292.1 PYGN01002371 PSN30636.1 CM000160 EDW99197.2 AE014297 AY052003 AAF57132.2 AAK93427.1 AFH06702.2 AFH06703.2 KRK04724.1 GBXI01006464 JAD07828.1 NEVH01005277 PNF39181.1 CH479186 EDW39182.1 CM000070 EAL26697.2 CH902617 EDV43300.2 KPU80190.1 EIM52265.2 CH954182 EDV52974.1 OUUW01000005 SPP80812.1 CH480819 EDW53611.1 CP012526 ALC48041.1 CM000364 EDX15103.1 SPP80814.1 KK852427 KDR24088.1 DS235745 EEB16231.1 CH964251 EDW82993.1 KRF99705.1 NNAY01002172 OXU21875.1 CH933806 EDW16262.2 CH940652 EDW59022.2 GEDC01031752 GEDC01012010 JAS05546.1 JAS25288.1 CH916377 EDV90802.1 GFPF01009225 MAA20371.1 GFAC01000790 JAT98398.1 GEDV01005093 JAP83464.1 GACK01008662 JAA56372.1 GBBM01006598 JAC28820.1 CP026243 AWO96737.1 FR905325 CDQ78399.1 DQ481664 ABF22395.1 GCES01010740 JAR75583.1 EF594311 ABS70749.1 AERX01033821 AYCK01013337
KPI99997.1 ODYU01006696 SOQ48770.1 AGBW02007989 OWR54349.1 GDHF01020486 JAI31828.1 GDHF01002932 JAI49382.1 GAKP01019566 JAC39386.1 GDHF01015022 JAI37292.1 PYGN01002371 PSN30636.1 CM000160 EDW99197.2 AE014297 AY052003 AAF57132.2 AAK93427.1 AFH06702.2 AFH06703.2 KRK04724.1 GBXI01006464 JAD07828.1 NEVH01005277 PNF39181.1 CH479186 EDW39182.1 CM000070 EAL26697.2 CH902617 EDV43300.2 KPU80190.1 EIM52265.2 CH954182 EDV52974.1 OUUW01000005 SPP80812.1 CH480819 EDW53611.1 CP012526 ALC48041.1 CM000364 EDX15103.1 SPP80814.1 KK852427 KDR24088.1 DS235745 EEB16231.1 CH964251 EDW82993.1 KRF99705.1 NNAY01002172 OXU21875.1 CH933806 EDW16262.2 CH940652 EDW59022.2 GEDC01031752 GEDC01012010 JAS05546.1 JAS25288.1 CH916377 EDV90802.1 GFPF01009225 MAA20371.1 GFAC01000790 JAT98398.1 GEDV01005093 JAP83464.1 GACK01008662 JAA56372.1 GBBM01006598 JAC28820.1 CP026243 AWO96737.1 FR905325 CDQ78399.1 DQ481664 ABF22395.1 GCES01010740 JAR75583.1 EF594311 ABS70749.1 AERX01033821 AYCK01013337
Proteomes
UP000005204
UP000053240
UP000218220
UP000053268
UP000007151
UP000245037
+ More
UP000002282 UP000000803 UP000235965 UP000008744 UP000001819 UP000192221 UP000007801 UP000008711 UP000268350 UP000001292 UP000092553 UP000000304 UP000027135 UP000009046 UP000007798 UP000215335 UP000002358 UP000009192 UP000008792 UP000001070 UP000092445 UP000265120 UP000246464 UP000193380 UP000087266 UP000261360 UP000261420 UP000007303 UP000005226 UP000265100 UP000265160 UP000261460 UP000264840 UP000005207 UP000261380 UP000028760
UP000002282 UP000000803 UP000235965 UP000008744 UP000001819 UP000192221 UP000007801 UP000008711 UP000268350 UP000001292 UP000092553 UP000000304 UP000027135 UP000009046 UP000007798 UP000215335 UP000002358 UP000009192 UP000008792 UP000001070 UP000092445 UP000265120 UP000246464 UP000193380 UP000087266 UP000261360 UP000261420 UP000007303 UP000005226 UP000265100 UP000265160 UP000261460 UP000264840 UP000005207 UP000261380 UP000028760
Pfam
PF03154 Atrophin-1
SUPFAM
SSF57667
SSF57667
ProteinModelPortal
H9JAC1
A0A194RAM9
A0A2A4J3T1
A0A194Q365
A0A2H1W6R9
A0A212FKY3
+ More
A0A0K8UZ63 A0A0K8WE69 A0A034V7V7 A0A0K8VEP6 A0A2P8XF54 B4PNB5 Q9V9Z6 A0A0B4K810 A0A0R1EBU8 A0A0A1XBI3 A0A2J7REB6 B4GN56 Q29CE9 A0A1W4UI25 A0A1W4UHZ4 B3M1K9 A0A0P9CA52 I5AMT0 B3P7R6 A0A3B0JKG7 B4I004 A0A0M4EHF6 B4QSK6 A0A3B0JEK7 A0A067RM36 E0VS75 B4NFC6 A0A0Q9X1F7 A0A232EU34 K7J2T0 B4K625 B4M5U6 A0A1B6BW75 B4JYP0 A0A1A9Z2E6 A0A224Z0D4 A0A1E1XGN9 A0A131YW98 L7LXQ2 A0A023G7G5 A0A3P8V531 A0A2U9AYQ9 A0A060XM74 A0A1S3M607 A0A3B4Y6A1 A0A3B4T5T0 A0A1S3Q1J8 H3DM52 Q1KKZ9 A0A147AAM3 A0A3P8NKD2 A0A3P9C9G1 A0A3B4GDS5 A8DSV7 I3J4A5 A0A3B5N0D4 A0A087XEK7
A0A0K8UZ63 A0A0K8WE69 A0A034V7V7 A0A0K8VEP6 A0A2P8XF54 B4PNB5 Q9V9Z6 A0A0B4K810 A0A0R1EBU8 A0A0A1XBI3 A0A2J7REB6 B4GN56 Q29CE9 A0A1W4UI25 A0A1W4UHZ4 B3M1K9 A0A0P9CA52 I5AMT0 B3P7R6 A0A3B0JKG7 B4I004 A0A0M4EHF6 B4QSK6 A0A3B0JEK7 A0A067RM36 E0VS75 B4NFC6 A0A0Q9X1F7 A0A232EU34 K7J2T0 B4K625 B4M5U6 A0A1B6BW75 B4JYP0 A0A1A9Z2E6 A0A224Z0D4 A0A1E1XGN9 A0A131YW98 L7LXQ2 A0A023G7G5 A0A3P8V531 A0A2U9AYQ9 A0A060XM74 A0A1S3M607 A0A3B4Y6A1 A0A3B4T5T0 A0A1S3Q1J8 H3DM52 Q1KKZ9 A0A147AAM3 A0A3P8NKD2 A0A3P9C9G1 A0A3B4GDS5 A8DSV7 I3J4A5 A0A3B5N0D4 A0A087XEK7
PDB
2YT9
E-value=0.0111503,
Score=90
Ontologies
GO
Topology
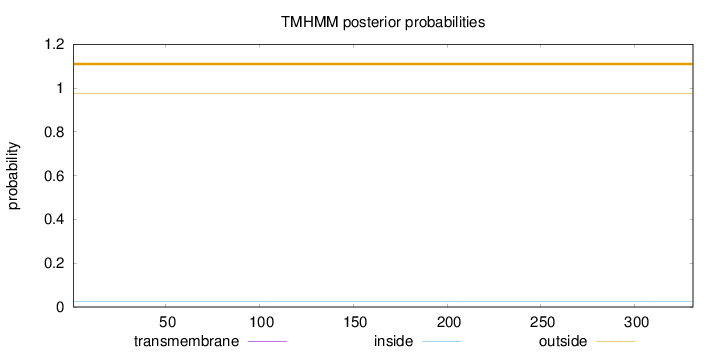
Length:
331
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000890000000000001
Exp number, first 60 AAs:
0.0006
Total prob of N-in:
0.02505
outside
1 - 331
Population Genetic Test Statistics
Pi
174.676526
Theta
145.154322
Tajima's D
0.548123
CLR
0
CSRT
0.527273636318184
Interpretation
Uncertain
