Pre Gene Modal
BGIBMGA006412
Annotation
PREDICTED:_ATP_synthase_lipid-binding_protein?_mitochondrial_[Bombyx_mori]
Full name
ATP synthase subunit c
+ More
ATP synthase lipid-binding protein, mitochondrial
ATP synthase lipid-binding protein, mitochondrial
Alternative Name
ATP synthase F(0) sector subunit c
F-type ATPase subunit c
Lipid-binding protein
ATPase protein 9
ATPase subunit c
F-type ATPase subunit c
Lipid-binding protein
ATPase protein 9
ATPase subunit c
Location in the cell
Mitochondrial Reliability : 1.753 PlasmaMembrane Reliability : 2.143
Sequence
CDS
ATGCTGTCTGCCGCCAGACTGATCGCCCCTGCAGCCAGGTCTGCCATCTTCTGCAACTCTGCACTGGTGCGACCACTTGCAGCAGTACCCACCCATACACAGATGGTACCTGCTGTCCCTACACAGCTCTCTGCAGTGCGGTCCTTCCAGACCACATCGGTCACTAAGGACATTGACTCTGCTGCCAAATTCATTGGTGCTGGTGCAGCGACAGTGGGAGTAGCTGGTTCCGGAGCTGGTATTGGAACAGTCTTCGGCTCCCTCATCATCGGCTATGCCAGGAACCCCTCCCTCAAGCAGCAGTTGTTCTCATACGCCATTCTGGGTTTCGCCTTGTCTGAGGCTATGGGTCTGTTCTGTCTTATGATGGCGTTCCTGCTGCTCTTCGCTTTCTAA
Protein
MLSAARLIAPAARSAIFCNSALVRPLAAVPTHTQMVPAVPTQLSAVRSFQTTSVTKDIDSAAKFIGAGAATVGVAGSGAGIGTVFGSLIIGYARNPSLKQQLFSYAILGFALSEAMGLFCLMMAFLLLFAF
Summary
Description
F(1)F(0) ATP synthase produces ATP from ADP in the presence of a proton or sodium gradient. F-type ATPases consist of two structural domains, F(1) containing the extramembraneous catalytic core and F(0) containing the membrane proton channel, linked together by a central stalk and a peripheral stalk. During catalysis, ATP synthesis in the catalytic domain of F(1) is coupled via a rotary mechanism of the central stalk subunits to proton translocation.
Key component of the F(0) channel; it plays a direct role in translocation across the membrane. A homomeric c-ring of between 10-14 subunits forms the central stalk rotor element with the F(1) delta and epsilon subunits.
Mitochondrial membrane ATP synthase (F(1)F(0) ATP synthase or Complex V) produces ATP from ADP in the presence of a proton gradient across the membrane which is generated by electron transport complexes of the respiratory chain. F-type ATPases consist of two structural domains, F(1) - containing the extramembraneous catalytic core and F(0) - containing the membrane proton channel, linked together by a central stalk and a peripheral stalk. During catalysis, ATP synthesis in the catalytic domain of F(1) is coupled via a rotary mechanism of the central stalk subunits to proton translocation. Part of the complex F(0) domain. A homomeric c-ring of probably 10 subunits is part of the complex rotary element (By similarity).
Key component of the F(0) channel; it plays a direct role in translocation across the membrane. A homomeric c-ring of between 10-14 subunits forms the central stalk rotor element with the F(1) delta and epsilon subunits.
Mitochondrial membrane ATP synthase (F(1)F(0) ATP synthase or Complex V) produces ATP from ADP in the presence of a proton gradient across the membrane which is generated by electron transport complexes of the respiratory chain. F-type ATPases consist of two structural domains, F(1) - containing the extramembraneous catalytic core and F(0) - containing the membrane proton channel, linked together by a central stalk and a peripheral stalk. During catalysis, ATP synthesis in the catalytic domain of F(1) is coupled via a rotary mechanism of the central stalk subunits to proton translocation. Part of the complex F(0) domain. A homomeric c-ring of probably 10 subunits is part of the complex rotary element (By similarity).
Subunit
F-type ATPases have 2 components, CF(1) - the catalytic core - and CF(0) - the membrane proton channel. CF(1) has five subunits: alpha(3), beta(3), gamma(1), delta(1), epsilon(1). CF(0) has three main subunits: a, b and c.
Similarity
Belongs to the ATPase C chain family.
Keywords
Hydrogen ion transport
Ion transport
Lipid-binding
Membrane
Methylation
Mitochondrion
Transit peptide
Transmembrane
Transmembrane helix
Transport
Feature
chain ATP synthase subunit c
Uniprot
A0A3G1T1C9
A0A3B0E4A8
Q9U505
A0A1E1WK31
A0A0L7LHM8
A0A076FLZ2
+ More
A0A2H1W6R3 I4DM35 I4DIK3 A0A212FKT3 A0A194R6P8 R4WHV9 A0A2P8XFA1 A0A3B4AN32 A0A067RJV5 C1BKB8 A0A1D1Y9C8 A0A0L7RB10 A0A3B4AMJ9 Q06DF7 A0A1I8JV93 A0A182IAB9 A0A182P7T5 A0A182LTA9 A0A182TS18 A0A182KYB2 A0A182XEF4 A7USF7 F1RWF6 A0A195DQM6 E2AC98 A0A3B5KMJ4 A0A023F9P0 A0A182YMX6 A0A182N078 A0A084VIW3 A0A1B0EY42 A0A0J7LBH3 A0A3P9AM30 C4N187 A0A182QH23 A0A1I8PHQ8 A0A3P9NDT9 A0A182J0U9 A0A182R9I8 A0A2U4AHQ6 A0A0S7GC10 A0A3P9NDT3 A0A3B3QNT0 A0A087XQH9 A0A2R7WMU1 A0A1A8R7Y3 A0A1A8KRI5 A0A1A8NUD0 A0A1A7YZA8 A0A1A8E919 A0A1A8GK12 A0A1A8AE94 A0A3B3Y0B6 M3ZDW0 A0A3B3UNB3 A0A060WWW1 B5DGN3 A0A151I5Z8 A0A158NAN0 F4W6W0 A0A195EWK9 A0A3B3U0T0 Q201X0 A0A1A9XUC5 A0A1A9UWX7 A0A1B0BHP6 A0A1B0AJW1 A0A1A9WZ22 D3TRM4 A0A158NAM9 T1DJ07 A0A1Q3FI93 B0WM99 Q1KKT5 B5XFQ8 Q29570 A0A1A8E7A0 A0A1A8FSE8 A0A1A8UWL5 A0A2M4A0W9 A0A1B6G2G8 A0A1S3A677 A0A182KA43 A0A3P8V4E0 A0A2U4AHM7 A0A2Y9LW16
A0A2H1W6R3 I4DM35 I4DIK3 A0A212FKT3 A0A194R6P8 R4WHV9 A0A2P8XFA1 A0A3B4AN32 A0A067RJV5 C1BKB8 A0A1D1Y9C8 A0A0L7RB10 A0A3B4AMJ9 Q06DF7 A0A1I8JV93 A0A182IAB9 A0A182P7T5 A0A182LTA9 A0A182TS18 A0A182KYB2 A0A182XEF4 A7USF7 F1RWF6 A0A195DQM6 E2AC98 A0A3B5KMJ4 A0A023F9P0 A0A182YMX6 A0A182N078 A0A084VIW3 A0A1B0EY42 A0A0J7LBH3 A0A3P9AM30 C4N187 A0A182QH23 A0A1I8PHQ8 A0A3P9NDT9 A0A182J0U9 A0A182R9I8 A0A2U4AHQ6 A0A0S7GC10 A0A3P9NDT3 A0A3B3QNT0 A0A087XQH9 A0A2R7WMU1 A0A1A8R7Y3 A0A1A8KRI5 A0A1A8NUD0 A0A1A7YZA8 A0A1A8E919 A0A1A8GK12 A0A1A8AE94 A0A3B3Y0B6 M3ZDW0 A0A3B3UNB3 A0A060WWW1 B5DGN3 A0A151I5Z8 A0A158NAN0 F4W6W0 A0A195EWK9 A0A3B3U0T0 Q201X0 A0A1A9XUC5 A0A1A9UWX7 A0A1B0BHP6 A0A1B0AJW1 A0A1A9WZ22 D3TRM4 A0A158NAM9 T1DJ07 A0A1Q3FI93 B0WM99 Q1KKT5 B5XFQ8 Q29570 A0A1A8E7A0 A0A1A8FSE8 A0A1A8UWL5 A0A2M4A0W9 A0A1B6G2G8 A0A1S3A677 A0A182KA43 A0A3P8V4E0 A0A2U4AHM7 A0A2Y9LW16
Pubmed
10620045
26227816
22651552
26354079
22118469
23691247
+ More
29403074 25463417 24845553 17244545 20966253 12364791 14747013 17210077 20798317 21551351 25474469 25244985 24438588 25069045 19576987 29240929 23542700 24755649 19878547 20433749 21347285 21719571 20353571 24330624 16636282 8672129 24487278
29403074 25463417 24845553 17244545 20966253 12364791 14747013 17210077 20798317 21551351 25474469 25244985 24438588 25069045 19576987 29240929 23542700 24755649 19878547 20433749 21347285 21719571 20353571 24330624 16636282 8672129 24487278
EMBL
MG846938
AXY94790.1
RBVL01000231
RKO10346.1
AF117583
GDQN01009827
+ More
GDQN01004041 GDQN01003684 GDQN01001743 GDQN01000082 JAT81227.1 JAT87013.1 JAT87370.1 JAT89311.1 JAT90972.1 JTDY01001076 KOB74957.1 KF803646 AII16888.1 ODYU01006696 SOQ48771.1 AK402353 BAM18975.1 AK401121 KQ459551 BAM17743.1 KPI99998.1 AGBW02007989 OWR54348.1 KQ460685 KPJ12915.1 AK416940 BAN20155.1 PYGN01002371 PSN30634.1 KK852427 KDR24087.1 BT075047 ACO09471.1 GDJX01016694 JAT51242.1 KQ414618 KOC67926.1 DQ910368 ABI83790.1 APCN01001140 AXCM01000565 AAAB01008846 EDO64318.1 AEMK02000080 KQ980612 KYN15168.1 GL438484 EFN68944.1 GBBI01001074 JAC17638.1 ATLV01013412 KE524855 KFB37907.1 AJVK01007418 LBMM01000031 KMR05262.1 EZ048860 ACN69152.1 AXCN02001257 GBYX01457045 GBYX01457044 JAO24499.1 AYCK01005506 KK855016 PTY20320.1 HAEH01014922 HAEI01016825 SBS01364.1 HAED01021143 HAEE01015045 SBR35095.1 HAEF01019196 HAEG01004852 SBR72661.1 HADW01011579 HADX01013271 SBP35503.1 HADZ01006361 HAEA01013114 SBQ41594.1 HAEB01017935 HAEC01003420 SBQ71497.1 HADY01014065 HAEJ01018056 SBP52550.1 FR904793 CDQ71838.1 BT043792 BT046489 BT058350 BT060050 ACH70907.1 ACI66290.1 ACN10063.1 ACN12410.1 KQ976413 KYM90349.1 ADTU01010396 GL887762 EGI70073.1 KQ981954 KYN32282.1 ABLF02033112 DQ413216 AK343131 ABD72684.1 BAH72902.1 JXJN01014481 EZ424076 EZ424078 ADD20352.1 ADD20354.1 GALA01000611 JAA94241.1 GFDL01007781 JAV27264.1 DS231996 EDS30948.1 DQ481668 ABF22459.1 BT049877 ACI69678.1 F14506 CAA23094.1 HADZ01004304 HAEA01012499 SBQ40979.1 HAEB01014527 HAEC01004864 SBQ61054.1 HADY01024320 HAEJ01011265 SBS51722.1 GGFK01001088 MBW34409.1 GECZ01026928 GECZ01015112 GECZ01013276 JAS42841.1 JAS54657.1 JAS56493.1
GDQN01004041 GDQN01003684 GDQN01001743 GDQN01000082 JAT81227.1 JAT87013.1 JAT87370.1 JAT89311.1 JAT90972.1 JTDY01001076 KOB74957.1 KF803646 AII16888.1 ODYU01006696 SOQ48771.1 AK402353 BAM18975.1 AK401121 KQ459551 BAM17743.1 KPI99998.1 AGBW02007989 OWR54348.1 KQ460685 KPJ12915.1 AK416940 BAN20155.1 PYGN01002371 PSN30634.1 KK852427 KDR24087.1 BT075047 ACO09471.1 GDJX01016694 JAT51242.1 KQ414618 KOC67926.1 DQ910368 ABI83790.1 APCN01001140 AXCM01000565 AAAB01008846 EDO64318.1 AEMK02000080 KQ980612 KYN15168.1 GL438484 EFN68944.1 GBBI01001074 JAC17638.1 ATLV01013412 KE524855 KFB37907.1 AJVK01007418 LBMM01000031 KMR05262.1 EZ048860 ACN69152.1 AXCN02001257 GBYX01457045 GBYX01457044 JAO24499.1 AYCK01005506 KK855016 PTY20320.1 HAEH01014922 HAEI01016825 SBS01364.1 HAED01021143 HAEE01015045 SBR35095.1 HAEF01019196 HAEG01004852 SBR72661.1 HADW01011579 HADX01013271 SBP35503.1 HADZ01006361 HAEA01013114 SBQ41594.1 HAEB01017935 HAEC01003420 SBQ71497.1 HADY01014065 HAEJ01018056 SBP52550.1 FR904793 CDQ71838.1 BT043792 BT046489 BT058350 BT060050 ACH70907.1 ACI66290.1 ACN10063.1 ACN12410.1 KQ976413 KYM90349.1 ADTU01010396 GL887762 EGI70073.1 KQ981954 KYN32282.1 ABLF02033112 DQ413216 AK343131 ABD72684.1 BAH72902.1 JXJN01014481 EZ424076 EZ424078 ADD20352.1 ADD20354.1 GALA01000611 JAA94241.1 GFDL01007781 JAV27264.1 DS231996 EDS30948.1 DQ481668 ABF22459.1 BT049877 ACI69678.1 F14506 CAA23094.1 HADZ01004304 HAEA01012499 SBQ40979.1 HAEB01014527 HAEC01004864 SBQ61054.1 HADY01024320 HAEJ01011265 SBS51722.1 GGFK01001088 MBW34409.1 GECZ01026928 GECZ01015112 GECZ01013276 JAS42841.1 JAS54657.1 JAS56493.1
Proteomes
UP000276569
UP000037510
UP000053268
UP000007151
UP000053240
UP000245037
+ More
UP000261520 UP000027135 UP000053825 UP000075903 UP000075840 UP000075885 UP000075883 UP000075902 UP000075882 UP000076407 UP000007062 UP000008227 UP000078492 UP000000311 UP000005226 UP000076408 UP000075884 UP000030765 UP000092462 UP000036403 UP000265140 UP000075886 UP000095300 UP000242638 UP000075880 UP000075900 UP000245320 UP000261540 UP000028760 UP000261480 UP000002852 UP000261500 UP000193380 UP000087266 UP000078540 UP000005205 UP000007755 UP000078541 UP000007819 UP000092443 UP000078200 UP000092460 UP000092445 UP000091820 UP000002320 UP000079721 UP000075881 UP000265120 UP000248483
UP000261520 UP000027135 UP000053825 UP000075903 UP000075840 UP000075885 UP000075883 UP000075902 UP000075882 UP000076407 UP000007062 UP000008227 UP000078492 UP000000311 UP000005226 UP000076408 UP000075884 UP000030765 UP000092462 UP000036403 UP000265140 UP000075886 UP000095300 UP000242638 UP000075880 UP000075900 UP000245320 UP000261540 UP000028760 UP000261480 UP000002852 UP000261500 UP000193380 UP000087266 UP000078540 UP000005205 UP000007755 UP000078541 UP000007819 UP000092443 UP000078200 UP000092460 UP000092445 UP000091820 UP000002320 UP000079721 UP000075881 UP000265120 UP000248483
PRIDE
Pfam
PF00137 ATP-synt_C
Interpro
SUPFAM
SSF81333
SSF81333
Gene 3D
ProteinModelPortal
A0A3G1T1C9
A0A3B0E4A8
Q9U505
A0A1E1WK31
A0A0L7LHM8
A0A076FLZ2
+ More
A0A2H1W6R3 I4DM35 I4DIK3 A0A212FKT3 A0A194R6P8 R4WHV9 A0A2P8XFA1 A0A3B4AN32 A0A067RJV5 C1BKB8 A0A1D1Y9C8 A0A0L7RB10 A0A3B4AMJ9 Q06DF7 A0A1I8JV93 A0A182IAB9 A0A182P7T5 A0A182LTA9 A0A182TS18 A0A182KYB2 A0A182XEF4 A7USF7 F1RWF6 A0A195DQM6 E2AC98 A0A3B5KMJ4 A0A023F9P0 A0A182YMX6 A0A182N078 A0A084VIW3 A0A1B0EY42 A0A0J7LBH3 A0A3P9AM30 C4N187 A0A182QH23 A0A1I8PHQ8 A0A3P9NDT9 A0A182J0U9 A0A182R9I8 A0A2U4AHQ6 A0A0S7GC10 A0A3P9NDT3 A0A3B3QNT0 A0A087XQH9 A0A2R7WMU1 A0A1A8R7Y3 A0A1A8KRI5 A0A1A8NUD0 A0A1A7YZA8 A0A1A8E919 A0A1A8GK12 A0A1A8AE94 A0A3B3Y0B6 M3ZDW0 A0A3B3UNB3 A0A060WWW1 B5DGN3 A0A151I5Z8 A0A158NAN0 F4W6W0 A0A195EWK9 A0A3B3U0T0 Q201X0 A0A1A9XUC5 A0A1A9UWX7 A0A1B0BHP6 A0A1B0AJW1 A0A1A9WZ22 D3TRM4 A0A158NAM9 T1DJ07 A0A1Q3FI93 B0WM99 Q1KKT5 B5XFQ8 Q29570 A0A1A8E7A0 A0A1A8FSE8 A0A1A8UWL5 A0A2M4A0W9 A0A1B6G2G8 A0A1S3A677 A0A182KA43 A0A3P8V4E0 A0A2U4AHM7 A0A2Y9LW16
A0A2H1W6R3 I4DM35 I4DIK3 A0A212FKT3 A0A194R6P8 R4WHV9 A0A2P8XFA1 A0A3B4AN32 A0A067RJV5 C1BKB8 A0A1D1Y9C8 A0A0L7RB10 A0A3B4AMJ9 Q06DF7 A0A1I8JV93 A0A182IAB9 A0A182P7T5 A0A182LTA9 A0A182TS18 A0A182KYB2 A0A182XEF4 A7USF7 F1RWF6 A0A195DQM6 E2AC98 A0A3B5KMJ4 A0A023F9P0 A0A182YMX6 A0A182N078 A0A084VIW3 A0A1B0EY42 A0A0J7LBH3 A0A3P9AM30 C4N187 A0A182QH23 A0A1I8PHQ8 A0A3P9NDT9 A0A182J0U9 A0A182R9I8 A0A2U4AHQ6 A0A0S7GC10 A0A3P9NDT3 A0A3B3QNT0 A0A087XQH9 A0A2R7WMU1 A0A1A8R7Y3 A0A1A8KRI5 A0A1A8NUD0 A0A1A7YZA8 A0A1A8E919 A0A1A8GK12 A0A1A8AE94 A0A3B3Y0B6 M3ZDW0 A0A3B3UNB3 A0A060WWW1 B5DGN3 A0A151I5Z8 A0A158NAN0 F4W6W0 A0A195EWK9 A0A3B3U0T0 Q201X0 A0A1A9XUC5 A0A1A9UWX7 A0A1B0BHP6 A0A1B0AJW1 A0A1A9WZ22 D3TRM4 A0A158NAM9 T1DJ07 A0A1Q3FI93 B0WM99 Q1KKT5 B5XFQ8 Q29570 A0A1A8E7A0 A0A1A8FSE8 A0A1A8UWL5 A0A2M4A0W9 A0A1B6G2G8 A0A1S3A677 A0A182KA43 A0A3P8V4E0 A0A2U4AHM7 A0A2Y9LW16
PDB
5FIL
E-value=8.74096e-11,
Score=154
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Cell membrane
Mitochondrion membrane
Mitochondrion membrane
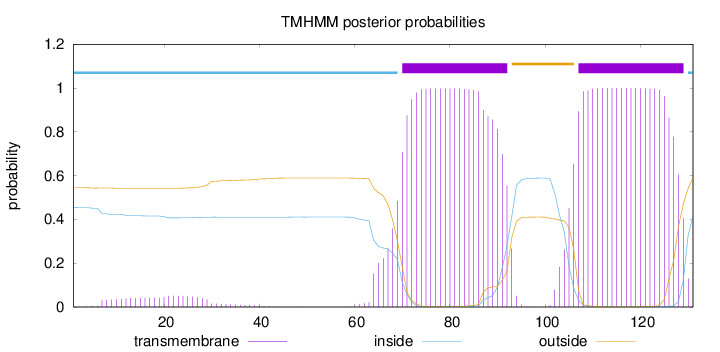
Length:
131
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
47.54873
Exp number, first 60 AAs:
1.11726
Total prob of N-in:
0.45406
inside
1 - 69
TMhelix
70 - 92
outside
93 - 106
TMhelix
107 - 129
inside
130 - 131
Population Genetic Test Statistics
Pi
252.497225
Theta
204.73786
Tajima's D
0.622887
CLR
0.059693
CSRT
0.560221988900555
Interpretation
Uncertain
