Gene
KWMTBOMO03313
Pre Gene Modal
BGIBMGA006464
Annotation
PREDICTED:_transmembrane_and_ubiquitin-like_domain-containing_protein_1_isoform_X1_[Bombyx_mori]
Full name
Angiotensin-converting enzyme
Location in the cell
Nuclear Reliability : 1.286
Sequence
CDS
ATGACCCTGATAGATGGGTTTGGTGATGAAGTAGTGCAGTTCATGGGGGTGGTCTTGACAGTTATTGTGGGAATATTGGCTTGGTGGTCAACCAATGCAAGACCTGATAGATACGGAACTGTACTAGTCCTGCGATCACGTCCAAACAGTCCTGTGGCAATTAGCATTCGACCCAGATCGCGACTGGCACTAGTGAATCAACAAGTACTTACAAATACTAGCCAAGAATCCAATGCTGATGCTGCAGGCACAAGCAGATCAGAAGTCCAAGAGACTCCCAGTGAAACTTCCAGGGTCATTCCCCTGCAGGAGATGGACAGCATAGTGGATGCCGATATGGCAATGCTTGATAACAACCGTTTACACTTTTACAGGAGAATGGACTCTCCAACAAACGCGCAAAGCAATCCAGAATCGACACAGGACGAGAGTGACGTACAGGAAGCAGCCGGGAATGCACAAATCAGGGAAATGGACAGCATAGTTAGCGCAATGGAGGCTGATGTCACTTCGGGATGTGATCTCTTTACGAGAGCACCGGTAGACGCAGCACAAGGGGACAGTGAAATGAAAAATGACAGTTCGAAAGACGGCGCTTCGTCTACCGATGTAGAGAATGCATCGGACGATGTGAACACATTAGTCCAAAATGCAGACACTCCCGCACTCCCGGAACGAGACGAGACAAAAAGAATCCTGATAAAACTGAAGTACCTAAACGACACCCTGAAGGAAGTTGACGGAAACCTTGATGAACTGTTAAAAGATTTTAAGTTACGTCATTTCTCAACGGAGCTGTCTTCCGAGTGCCGGATACGTCTGATATTCAACGGGCGCGTGCTGGCGGACGACGCGGCCACGCTGCAGGCGTGCGGGCTGCACGACCGCGCCGTCGTGCACTGCCTCGTGCATCCCAAGCGAGCGGGGCCGGGTGCGGCTGAGGAGAGCAGCGCGGCGGCCACGCACGAGCTGATCACGGAGAGGCCGCAGCCCGAGAGGTCTTGGGATTTAGAAAACATTCTGATGACGTTTGTCTCTGTCGCCCTCACCGTTGTATGGTTCTTTAGGTGTGAGTACTCAAACATGTTCACCGCAAGTGCTAGTGTAGCATTGTTCGGTCTGACGGTGTTCTACAGTGTTGCCATATTCGGCCTCTATCTCTCCGACACATTCCAGTTCGACCGACGACACCAGCCGCCAACGACGAACAACTGA
Protein
MTLIDGFGDEVVQFMGVVLTVIVGILAWWSTNARPDRYGTVLVLRSRPNSPVAISIRPRSRLALVNQQVLTNTSQESNADAAGTSRSEVQETPSETSRVIPLQEMDSIVDADMAMLDNNRLHFYRRMDSPTNAQSNPESTQDESDVQEAAGNAQIREMDSIVSAMEADVTSGCDLFTRAPVDAAQGDSEMKNDSSKDGASSTDVENASDDVNTLVQNADTPALPERDETKRILIKLKYLNDTLKEVDGNLDELLKDFKLRHFSTELSSECRIRLIFNGRVLADDAATLQACGLHDRAVVHCLVHPKRAGPGAAEESSAAATHELITERPQPERSWDLENILMTFVSVALTVVWFFRCEYSNMFTASASVALFGLTVFYSVAIFGLYLSDTFQFDRRHQPPTTNN
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M2 family.
Uniprot
H9JAB9
A0A2H1VVS2
A0A194Q370
A0A2W1BTK9
A0A212FNA9
A0A194R6P3
+ More
A0A1E1VYK2 A0A1E1WG28 A0A1E1WA99 A0A067RHA8 A0A1W4WU16 A0A1Y1KLD0 E0VMA3 A0A2J7PWN2 A0A1B6K4R4 U4USC9 A0A1B6CL94 A0A224XKQ3 A0A1B6E715 A0A2A3EBN8 V9IIE6 A0A026W5S1 A0A158NRA2 A0A0L7RG79 A0A195BEA7 A0A151WN54 F4WB08 V5H1N7 E2A0G0 A0A195FJJ9 A0A146LYJ8 A0A0A9YWL6 A0A0K8SZ73 E2BLD1 E9IRZ1 A0A151IFM2 T1ISP9 A0A0K8TPK1 A0A0T6B465 A0A336LTT3 K1P6V6 A0A1L8DSK2 A0A1L8DSQ5 A0A1Q3FB22 A0A1B0D302 A0A088AN85 U5ENK6 A0A182XFI5 A0A182I811 B0WGQ9 A0A023GH81 G3MKG8 A0A182UNX3 A0A182M621
A0A1E1VYK2 A0A1E1WG28 A0A1E1WA99 A0A067RHA8 A0A1W4WU16 A0A1Y1KLD0 E0VMA3 A0A2J7PWN2 A0A1B6K4R4 U4USC9 A0A1B6CL94 A0A224XKQ3 A0A1B6E715 A0A2A3EBN8 V9IIE6 A0A026W5S1 A0A158NRA2 A0A0L7RG79 A0A195BEA7 A0A151WN54 F4WB08 V5H1N7 E2A0G0 A0A195FJJ9 A0A146LYJ8 A0A0A9YWL6 A0A0K8SZ73 E2BLD1 E9IRZ1 A0A151IFM2 T1ISP9 A0A0K8TPK1 A0A0T6B465 A0A336LTT3 K1P6V6 A0A1L8DSK2 A0A1L8DSQ5 A0A1Q3FB22 A0A1B0D302 A0A088AN85 U5ENK6 A0A182XFI5 A0A182I811 B0WGQ9 A0A023GH81 G3MKG8 A0A182UNX3 A0A182M621
EC Number
3.4.-.-
Pubmed
EMBL
BABH01009008
BABH01009009
ODYU01004739
SOQ44935.1
KQ459551
KPJ00003.1
+ More
KZ149939 PZC76964.1 AGBW02006689 OWR55203.1 KQ460685 KPJ12910.1 GDQN01011247 JAT79807.1 GDQN01005125 JAT85929.1 GDQN01007263 JAT83791.1 KK852470 KDR23231.1 GEZM01080331 JAV62213.1 DS235305 EEB14509.1 NEVH01020869 PNF20757.1 GECU01004366 GECU01001312 JAT03341.1 JAT06395.1 KB632345 ERL93081.1 GEDC01023135 JAS14163.1 GFTR01004782 JAW11644.1 GEDC01014363 GEDC01003568 JAS22935.1 JAS33730.1 KZ288292 PBC29145.1 JR047438 AEY60417.1 KK107390 QOIP01000007 EZA51422.1 RLU20426.1 ADTU01023838 KQ414596 KOC69967.1 KQ976504 KYM82903.1 KQ982922 KYQ49250.1 GL888056 EGI68628.1 GALX01001728 JAB66738.1 GL435586 EFN73097.1 KQ981522 KYN40578.1 GDHC01006594 JAQ12035.1 GBHO01008118 GBHO01008116 JAG35486.1 JAG35488.1 GBRD01007249 JAG58572.1 GL449017 EFN83483.1 GL765283 EFZ16608.1 KQ977771 KYM99998.1 JH431446 GDAI01001537 JAI16066.1 LJIG01016015 KRT81869.1 UFQS01000188 UFQT01000188 SSX00980.1 SSX21360.1 JH819192 EKC19372.1 GFDF01004703 JAV09381.1 GFDF01004704 JAV09380.1 GFDL01010298 JAV24747.1 AJVK01010808 AJVK01010809 GANO01004037 JAB55834.1 APCN01002607 DS231928 EDS27135.1 GBBM01003348 JAC32070.1 JO842369 AEO33986.1 AXCM01000405
KZ149939 PZC76964.1 AGBW02006689 OWR55203.1 KQ460685 KPJ12910.1 GDQN01011247 JAT79807.1 GDQN01005125 JAT85929.1 GDQN01007263 JAT83791.1 KK852470 KDR23231.1 GEZM01080331 JAV62213.1 DS235305 EEB14509.1 NEVH01020869 PNF20757.1 GECU01004366 GECU01001312 JAT03341.1 JAT06395.1 KB632345 ERL93081.1 GEDC01023135 JAS14163.1 GFTR01004782 JAW11644.1 GEDC01014363 GEDC01003568 JAS22935.1 JAS33730.1 KZ288292 PBC29145.1 JR047438 AEY60417.1 KK107390 QOIP01000007 EZA51422.1 RLU20426.1 ADTU01023838 KQ414596 KOC69967.1 KQ976504 KYM82903.1 KQ982922 KYQ49250.1 GL888056 EGI68628.1 GALX01001728 JAB66738.1 GL435586 EFN73097.1 KQ981522 KYN40578.1 GDHC01006594 JAQ12035.1 GBHO01008118 GBHO01008116 JAG35486.1 JAG35488.1 GBRD01007249 JAG58572.1 GL449017 EFN83483.1 GL765283 EFZ16608.1 KQ977771 KYM99998.1 JH431446 GDAI01001537 JAI16066.1 LJIG01016015 KRT81869.1 UFQS01000188 UFQT01000188 SSX00980.1 SSX21360.1 JH819192 EKC19372.1 GFDF01004703 JAV09381.1 GFDF01004704 JAV09380.1 GFDL01010298 JAV24747.1 AJVK01010808 AJVK01010809 GANO01004037 JAB55834.1 APCN01002607 DS231928 EDS27135.1 GBBM01003348 JAC32070.1 JO842369 AEO33986.1 AXCM01000405
Proteomes
UP000005204
UP000053268
UP000007151
UP000053240
UP000027135
UP000192223
+ More
UP000009046 UP000235965 UP000030742 UP000242457 UP000053097 UP000279307 UP000005205 UP000053825 UP000078540 UP000075809 UP000007755 UP000000311 UP000078541 UP000008237 UP000078542 UP000005408 UP000092462 UP000005203 UP000076407 UP000075840 UP000002320 UP000075903 UP000075883
UP000009046 UP000235965 UP000030742 UP000242457 UP000053097 UP000279307 UP000005205 UP000053825 UP000078540 UP000075809 UP000007755 UP000000311 UP000078541 UP000008237 UP000078542 UP000005408 UP000092462 UP000005203 UP000076407 UP000075840 UP000002320 UP000075903 UP000075883
PRIDE
Interpro
SUPFAM
SSF54236
SSF54236
ProteinModelPortal
H9JAB9
A0A2H1VVS2
A0A194Q370
A0A2W1BTK9
A0A212FNA9
A0A194R6P3
+ More
A0A1E1VYK2 A0A1E1WG28 A0A1E1WA99 A0A067RHA8 A0A1W4WU16 A0A1Y1KLD0 E0VMA3 A0A2J7PWN2 A0A1B6K4R4 U4USC9 A0A1B6CL94 A0A224XKQ3 A0A1B6E715 A0A2A3EBN8 V9IIE6 A0A026W5S1 A0A158NRA2 A0A0L7RG79 A0A195BEA7 A0A151WN54 F4WB08 V5H1N7 E2A0G0 A0A195FJJ9 A0A146LYJ8 A0A0A9YWL6 A0A0K8SZ73 E2BLD1 E9IRZ1 A0A151IFM2 T1ISP9 A0A0K8TPK1 A0A0T6B465 A0A336LTT3 K1P6V6 A0A1L8DSK2 A0A1L8DSQ5 A0A1Q3FB22 A0A1B0D302 A0A088AN85 U5ENK6 A0A182XFI5 A0A182I811 B0WGQ9 A0A023GH81 G3MKG8 A0A182UNX3 A0A182M621
A0A1E1VYK2 A0A1E1WG28 A0A1E1WA99 A0A067RHA8 A0A1W4WU16 A0A1Y1KLD0 E0VMA3 A0A2J7PWN2 A0A1B6K4R4 U4USC9 A0A1B6CL94 A0A224XKQ3 A0A1B6E715 A0A2A3EBN8 V9IIE6 A0A026W5S1 A0A158NRA2 A0A0L7RG79 A0A195BEA7 A0A151WN54 F4WB08 V5H1N7 E2A0G0 A0A195FJJ9 A0A146LYJ8 A0A0A9YWL6 A0A0K8SZ73 E2BLD1 E9IRZ1 A0A151IFM2 T1ISP9 A0A0K8TPK1 A0A0T6B465 A0A336LTT3 K1P6V6 A0A1L8DSK2 A0A1L8DSQ5 A0A1Q3FB22 A0A1B0D302 A0A088AN85 U5ENK6 A0A182XFI5 A0A182I811 B0WGQ9 A0A023GH81 G3MKG8 A0A182UNX3 A0A182M621
PDB
1WIA
E-value=0.000756626,
Score=101
Ontologies
GO
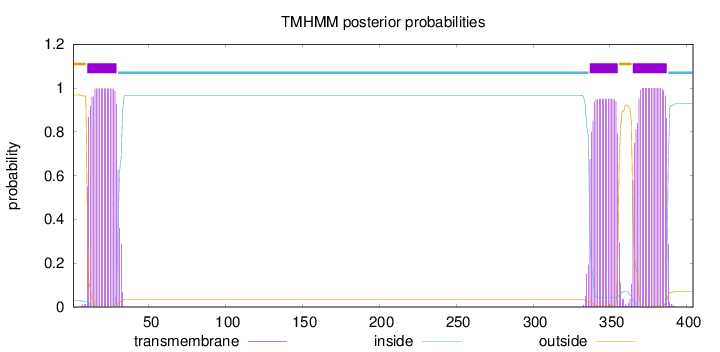
Topology
Length:
404
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
61.00569
Exp number, first 60 AAs:
20.52797
Total prob of N-in:
0.03126
POSSIBLE N-term signal
sequence
outside
1 - 9
TMhelix
10 - 29
inside
30 - 336
TMhelix
337 - 355
outside
356 - 364
TMhelix
365 - 387
inside
388 - 404
Population Genetic Test Statistics
Pi
217.36407
Theta
179.713626
Tajima's D
0.534778
CLR
0.024508
CSRT
0.530023498825059
Interpretation
Uncertain
