Gene
KWMTBOMO03310
Pre Gene Modal
BGIBMGA006417
Annotation
PREDICTED:_tyrosine-protein_kinase_Fps85D_isoform_X2_[Amyelois_transitella]
Full name
Tyrosine-protein kinase
+ More
Tyrosine-protein kinase Fer
Non-specific protein-tyrosine kinase
Tyrosine-protein kinase Fer
Non-specific protein-tyrosine kinase
Location in the cell
Cytoplasmic Reliability : 1.655 Nuclear Reliability : 1.947
Sequence
CDS
ATGGGATACACTACGAATGGCGCGGGTCGGGCAGCGCACGAGGCGCTGTTGGCGCGACAGGATGCTGAGCTAAGGCTCATGGAAGCTATGAAGAGAAGCCTCCAAGCAAAGATGAAAAGCGACAGAGAATATGCTCTGGCGCTAAGTGCTGCGGCTGCGCAGGGGCAGAAGATGGACAAATGCGAAGAGCTAAATGGATCGGTGATAGCGGCAGCATGGCGTACCATGACCGAGGAGTGGGATAACACAAGTCGTCTCATCAAATTGAATGCCGAAGCTCTCGAGTCTCGAGCTCTTGATCGCCTCACCACACTGATGACAGAACGAAGGAAGGCGCGCAAAATGTACCAGGAGGATCATACAAAAATCTCTTCTCAGTTCACACAGCTATCCGAAGAAGTGCAGAGAAAACAGAGCGAGTACCAAAAGTGCCTTGAGTCTTACAAACAGATGCGGGCCAAGTTCGAAGAGAATTATCTCAAATGTCCAACTAGCAGGGGAGGGAGGAAATTAGATGAGACGAGAGACAAATATGGCAAAGCGTGCAGGAGACTCCATCGGGCACACAACGAGTATGTGTTATTGTTGGCTGAGGCGACCGAATGCGAACGAGCCCTTCGGACTGCAGCCATCCCTGCGCTTTTAGAACAGCAACGTCGCCAGGGGGAAGCTACTGCTACTTCTTGGAAAGGGATCCTTTTAGAAGTGGTGCAGCGGTCAGACTATAGTACCGAAAAGTTCAGGGAGATCCAGACCAAAGTCGAGACTACAGTGCAGAATCTGAGGCCACAAGACGAGTACAAAGAGTTCTTAGAGAAGTATAAATCGCCTCCATCGACACCAGTGATATTTACATTTGACGAAAGCCTAGTGGAAGACACGAGCGGTAAATTACAACCAAATCAACTGACAGTCGATAATCTCACAGTTGAGTGGCTGAAAGAGAAACTCACAGAGCTGGAGGCAACATTGCAGGACAACAGAGAGAAACAGACCGCTTTGCAGGGACCAGAAATTATAGGGAATGGGGTTTGCAAAAACAACAATACCGGACTCACGAACGGCGTAAATGGAATCGACAGGCATTCTCCTCCGCCGATTCCCGTCACAACATCGGAGCCCATAAAGTTGTTAGAGCTACGCGTAGCAGAACGCAAATGTGCGAAACAAGCGGAGATGATACGATCCGCGTTGGCCGAACTAGGATGTGAAGAAGCGCCCAGCGGCTGCCCGGATCTGTCGGCCGAGACAGTCGGCGTTGATAGCTTAGACGGAAGCTCTCCCGATCATCAGGTGAGCGTGAGTGAGACAGAGCGTTCTCTAGTGGAACAGGAGTGGTTCCACGGTGTATTACCACGCGAGGAGGTGGTGAGGCTGTTGCGTAGTGACGGAGACTACCTAGTCCGTGAAACAACCAGGAACCACGCGAGGCAGCTAGTGTTATCCGTTTGCTGGGGACAGCATAAGCACTTCATTGTACAGACCACGCCAGAGGGTCACTATCGTTTCGAAGGAGCAGCGTTCCCGTCCGTGGCCGAGCTGATAGCGTGGCAGAGGGCTAGCGGCGTGCCCGTGACGGCGCGTTCGGGCGCGTTACTGCGGCGCGCGGTGCCCAGAGAGAACTGGGAACTCAACAACGATGATGTACAACTGCTCGATAAGATTGGAAGGGGTAATTTCGGTGACGTGTATAAAGCGCGTCTGAAGACGACAGGACAGGAGGTAGCCGTGAAGACGTGTAGGGTTGCGCTACCAGAGGAGCAGAAGCGGACGTTCCTACAGGAGGGACGGATCCTCAAGCAGTACCAGCACCCTAACATCGTCCGGCTTATTGGCATCGCCGTACAGAAGCAACCCATTATGATTGTCATGGAGCTCGTTTCAGGTACAAATTATTATAATTTCCAGAATAATATTATGATTTTTATTGCTTACTAG
Protein
MGYTTNGAGRAAHEALLARQDAELRLMEAMKRSLQAKMKSDREYALALSAAAAQGQKMDKCEELNGSVIAAAWRTMTEEWDNTSRLIKLNAEALESRALDRLTTLMTERRKARKMYQEDHTKISSQFTQLSEEVQRKQSEYQKCLESYKQMRAKFEENYLKCPTSRGGRKLDETRDKYGKACRRLHRAHNEYVLLLAEATECERALRTAAIPALLEQQRRQGEATATSWKGILLEVVQRSDYSTEKFREIQTKVETTVQNLRPQDEYKEFLEKYKSPPSTPVIFTFDESLVEDTSGKLQPNQLTVDNLTVEWLKEKLTELEATLQDNREKQTALQGPEIIGNGVCKNNNTGLTNGVNGIDRHSPPPIPVTTSEPIKLLELRVAERKCAKQAEMIRSALAELGCEEAPSGCPDLSAETVGVDSLDGSSPDHQVSVSETERSLVEQEWFHGVLPREEVVRLLRSDGDYLVRETTRNHARQLVLSVCWGQHKHFIVQTTPEGHYRFEGAAFPSVAELIAWQRASGVPVTARSGALLRRAVPRENWELNNDDVQLLDKIGRGNFGDVYKARLKTTGQEVAVKTCRVALPEEQKRTFLQEGRILKQYQHPNIVRLIGIAVQKQPIMIVMELVSGTNYYNFQNNIMIFIAY
Summary
Description
Tyrosine-protein kinase which is required during embryogenesis for formation of the actin cable in leading edge cells of the dorsal epidermis and for the timely progression of dorsal closure. May play a role in regulation of adherens junctions and cell adhesion through phosphorylation of the beta-catenin arm.
Catalytic Activity
ATP + L-tyrosyl-[protein] = ADP + H(+) + O-phospho-L-tyrosyl-[protein]
Similarity
Belongs to the protein kinase superfamily. Tyr protein kinase family. Fes/fps subfamily.
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Keywords
Alternative splicing
ATP-binding
Cell junction
Coiled coil
Complete proteome
Developmental protein
Kinase
Membrane
Nucleotide-binding
Phosphoprotein
Reference proteome
SH2 domain
Transferase
Tyrosine-protein kinase
Feature
chain Tyrosine-protein kinase Fer
splice variant In isoform D.
splice variant In isoform D.
Uniprot
A0A1W4X589
A0A139WAL6
A0A139WAJ6
A0A2J7REC4
A0A212FNB9
A0A139WAM2
+ More
A0A1W4X422 A0A1Y1KG60 A0A1Y1KFP9 U5EPN0 E2C012 A0A1L8DLX7 A0A0Q9X890 A0A026VZI7 A0A034V6J5 A0A2C9H8G3 W8ATK3 A0A2C9GPE0 A0A0Q9WMK3 A0A0K8UB42 K7IYX7 A0A182FZQ9 A0A1Q3FTM0 A0A0Q9X355 A0A0R1E298 P18106-1 A0A1W4VQ84 A0A3B0K594 A0A0R3NKN3 A0A0P9APC1 A0A0P8XY13 A0A087ZYS3 A0A151WIH7 A0A1B6F723 A0A0L7RDA6 A0A023F4U3 A0A1I8Q228 F5HLC9 A0A0A9VXQ2 A0A0A9W0J5 E0VKB1 A0A087UB48 A0A224XJB3 T1J3P8 A0A146KSL8 A0A1J1J7X7 A0A1B6CBF0 A0A146KYY4 A0A0P4Z062 A0A0P5FXE6 A0A0P5STU2 A0A0J7KUC9 A0A0M5J2E4 A0A0Q9WXK2 A0A0R1E1X2 A0A0B4KGK6 A0A0Q9WR25 A0A0R1E1T7 A0A0Q9WTA4 A0A0B4KFK7 B4K4Y8 B4HKF0 A0A1W4W2B3 B4PUM4 P18106 A0A1B6FKQ6 B3P1Q9
A0A1W4X422 A0A1Y1KG60 A0A1Y1KFP9 U5EPN0 E2C012 A0A1L8DLX7 A0A0Q9X890 A0A026VZI7 A0A034V6J5 A0A2C9H8G3 W8ATK3 A0A2C9GPE0 A0A0Q9WMK3 A0A0K8UB42 K7IYX7 A0A182FZQ9 A0A1Q3FTM0 A0A0Q9X355 A0A0R1E298 P18106-1 A0A1W4VQ84 A0A3B0K594 A0A0R3NKN3 A0A0P9APC1 A0A0P8XY13 A0A087ZYS3 A0A151WIH7 A0A1B6F723 A0A0L7RDA6 A0A023F4U3 A0A1I8Q228 F5HLC9 A0A0A9VXQ2 A0A0A9W0J5 E0VKB1 A0A087UB48 A0A224XJB3 T1J3P8 A0A146KSL8 A0A1J1J7X7 A0A1B6CBF0 A0A146KYY4 A0A0P4Z062 A0A0P5FXE6 A0A0P5STU2 A0A0J7KUC9 A0A0M5J2E4 A0A0Q9WXK2 A0A0R1E1X2 A0A0B4KGK6 A0A0Q9WR25 A0A0R1E1T7 A0A0Q9WTA4 A0A0B4KFK7 B4K4Y8 B4HKF0 A0A1W4W2B3 B4PUM4 P18106 A0A1B6FKQ6 B3P1Q9
EC Number
2.7.10.2
Pubmed
EMBL
KQ971382
KYB24939.1
KYB24938.1
NEVH01005277
PNF39177.1
AGBW02006689
+ More
OWR55199.1 KYB24940.1 GEZM01084983 JAV60344.1 GEZM01084984 JAV60343.1 GANO01004628 JAB55243.1 GL451712 EFN78708.1 GFDF01006625 JAV07459.1 CH933806 KRG00743.1 KK107652 EZA48284.1 GAKP01020013 GAKP01020011 JAC38941.1 GAMC01014380 JAB92175.1 APCN01003223 APCN01003224 APCN01003225 APCN01003226 APCN01003227 APCN01003228 APCN01003229 CH940650 KRF82737.1 GDHF01028521 JAI23793.1 GFDL01004084 JAV30961.1 CH964232 KRF99341.1 CM000160 KRK03253.1 X52844 U50450 AE014297 BT003462 AJ002916 OUUW01000005 SPP81169.1 CM000070 KRS99698.1 CH902617 KPU79527.1 KPU79526.1 KQ983089 KYQ47663.1 GECZ01023737 JAS46032.1 KQ414613 KOC68952.1 GBBI01002679 JAC16033.1 AAAB01008888 EGK97090.1 GBHO01043648 GBRD01006599 JAF99955.1 JAG59222.1 GBHO01043646 GBRD01006598 JAF99957.1 JAG59223.1 DS235241 EEB13817.1 KK119063 KFM74587.1 GFTR01007876 JAW08550.1 JH431830 GDHC01020422 JAP98206.1 CVRI01000074 CRL07876.1 GEDC01026629 JAS10669.1 GDHC01018293 JAQ00336.1 GDIP01219564 JAJ03838.1 GDIQ01250321 JAK01404.1 GDIP01140633 JAL63081.1 LBMM01003172 KMQ93859.1 CP012526 ALC47136.1 KRG00742.1 KRK03256.1 AGB95793.1 KRF82736.1 KRK03254.1 KRF99340.1 AGB95794.1 EDW13959.2 CH480815 EDW42900.1 EDW96641.1 GECZ01019008 JAS50761.1 CH954181 EDV49797.1
OWR55199.1 KYB24940.1 GEZM01084983 JAV60344.1 GEZM01084984 JAV60343.1 GANO01004628 JAB55243.1 GL451712 EFN78708.1 GFDF01006625 JAV07459.1 CH933806 KRG00743.1 KK107652 EZA48284.1 GAKP01020013 GAKP01020011 JAC38941.1 GAMC01014380 JAB92175.1 APCN01003223 APCN01003224 APCN01003225 APCN01003226 APCN01003227 APCN01003228 APCN01003229 CH940650 KRF82737.1 GDHF01028521 JAI23793.1 GFDL01004084 JAV30961.1 CH964232 KRF99341.1 CM000160 KRK03253.1 X52844 U50450 AE014297 BT003462 AJ002916 OUUW01000005 SPP81169.1 CM000070 KRS99698.1 CH902617 KPU79527.1 KPU79526.1 KQ983089 KYQ47663.1 GECZ01023737 JAS46032.1 KQ414613 KOC68952.1 GBBI01002679 JAC16033.1 AAAB01008888 EGK97090.1 GBHO01043648 GBRD01006599 JAF99955.1 JAG59222.1 GBHO01043646 GBRD01006598 JAF99957.1 JAG59223.1 DS235241 EEB13817.1 KK119063 KFM74587.1 GFTR01007876 JAW08550.1 JH431830 GDHC01020422 JAP98206.1 CVRI01000074 CRL07876.1 GEDC01026629 JAS10669.1 GDHC01018293 JAQ00336.1 GDIP01219564 JAJ03838.1 GDIQ01250321 JAK01404.1 GDIP01140633 JAL63081.1 LBMM01003172 KMQ93859.1 CP012526 ALC47136.1 KRG00742.1 KRK03256.1 AGB95793.1 KRF82736.1 KRK03254.1 KRF99340.1 AGB95794.1 EDW13959.2 CH480815 EDW42900.1 EDW96641.1 GECZ01019008 JAS50761.1 CH954181 EDV49797.1
Proteomes
UP000192223
UP000007266
UP000235965
UP000007151
UP000008237
UP000009192
+ More
UP000053097 UP000076407 UP000075840 UP000008792 UP000002358 UP000069272 UP000007798 UP000002282 UP000000803 UP000192221 UP000268350 UP000001819 UP000007801 UP000005203 UP000075809 UP000053825 UP000095300 UP000007062 UP000009046 UP000054359 UP000183832 UP000036403 UP000092553 UP000001292 UP000008711
UP000053097 UP000076407 UP000075840 UP000008792 UP000002358 UP000069272 UP000007798 UP000002282 UP000000803 UP000192221 UP000268350 UP000001819 UP000007801 UP000005203 UP000075809 UP000053825 UP000095300 UP000007062 UP000009046 UP000054359 UP000183832 UP000036403 UP000092553 UP000001292 UP000008711
PRIDE
Interpro
IPR001245
Ser-Thr/Tyr_kinase_cat_dom
+ More
IPR011009 Kinase-like_dom_sf
IPR035849 Fes/Fps/Fer_SH2
IPR017441 Protein_kinase_ATP_BS
IPR008266 Tyr_kinase_AS
IPR016250 Tyr-prot_kinase_Fes/Fps
IPR027267 AH/BAR_dom_sf
IPR020635 Tyr_kinase_cat_dom
IPR001060 FCH_dom
IPR036860 SH2_dom_sf
IPR000980 SH2
IPR031160 F_BAR
IPR000719 Prot_kinase_dom
IPR003604 Matrin/U1-like-C_Znf_C2H2
IPR013087 Znf_C2H2_type
IPR006561 DZF_dom
IPR036236 Znf_C2H2_sf
IPR028539 Fer
IPR011009 Kinase-like_dom_sf
IPR035849 Fes/Fps/Fer_SH2
IPR017441 Protein_kinase_ATP_BS
IPR008266 Tyr_kinase_AS
IPR016250 Tyr-prot_kinase_Fes/Fps
IPR027267 AH/BAR_dom_sf
IPR020635 Tyr_kinase_cat_dom
IPR001060 FCH_dom
IPR036860 SH2_dom_sf
IPR000980 SH2
IPR031160 F_BAR
IPR000719 Prot_kinase_dom
IPR003604 Matrin/U1-like-C_Znf_C2H2
IPR013087 Znf_C2H2_type
IPR006561 DZF_dom
IPR036236 Znf_C2H2_sf
IPR028539 Fer
Gene 3D
ProteinModelPortal
A0A1W4X589
A0A139WAL6
A0A139WAJ6
A0A2J7REC4
A0A212FNB9
A0A139WAM2
+ More
A0A1W4X422 A0A1Y1KG60 A0A1Y1KFP9 U5EPN0 E2C012 A0A1L8DLX7 A0A0Q9X890 A0A026VZI7 A0A034V6J5 A0A2C9H8G3 W8ATK3 A0A2C9GPE0 A0A0Q9WMK3 A0A0K8UB42 K7IYX7 A0A182FZQ9 A0A1Q3FTM0 A0A0Q9X355 A0A0R1E298 P18106-1 A0A1W4VQ84 A0A3B0K594 A0A0R3NKN3 A0A0P9APC1 A0A0P8XY13 A0A087ZYS3 A0A151WIH7 A0A1B6F723 A0A0L7RDA6 A0A023F4U3 A0A1I8Q228 F5HLC9 A0A0A9VXQ2 A0A0A9W0J5 E0VKB1 A0A087UB48 A0A224XJB3 T1J3P8 A0A146KSL8 A0A1J1J7X7 A0A1B6CBF0 A0A146KYY4 A0A0P4Z062 A0A0P5FXE6 A0A0P5STU2 A0A0J7KUC9 A0A0M5J2E4 A0A0Q9WXK2 A0A0R1E1X2 A0A0B4KGK6 A0A0Q9WR25 A0A0R1E1T7 A0A0Q9WTA4 A0A0B4KFK7 B4K4Y8 B4HKF0 A0A1W4W2B3 B4PUM4 P18106 A0A1B6FKQ6 B3P1Q9
A0A1W4X422 A0A1Y1KG60 A0A1Y1KFP9 U5EPN0 E2C012 A0A1L8DLX7 A0A0Q9X890 A0A026VZI7 A0A034V6J5 A0A2C9H8G3 W8ATK3 A0A2C9GPE0 A0A0Q9WMK3 A0A0K8UB42 K7IYX7 A0A182FZQ9 A0A1Q3FTM0 A0A0Q9X355 A0A0R1E298 P18106-1 A0A1W4VQ84 A0A3B0K594 A0A0R3NKN3 A0A0P9APC1 A0A0P8XY13 A0A087ZYS3 A0A151WIH7 A0A1B6F723 A0A0L7RDA6 A0A023F4U3 A0A1I8Q228 F5HLC9 A0A0A9VXQ2 A0A0A9W0J5 E0VKB1 A0A087UB48 A0A224XJB3 T1J3P8 A0A146KSL8 A0A1J1J7X7 A0A1B6CBF0 A0A146KYY4 A0A0P4Z062 A0A0P5FXE6 A0A0P5STU2 A0A0J7KUC9 A0A0M5J2E4 A0A0Q9WXK2 A0A0R1E1X2 A0A0B4KGK6 A0A0Q9WR25 A0A0R1E1T7 A0A0Q9WTA4 A0A0B4KFK7 B4K4Y8 B4HKF0 A0A1W4W2B3 B4PUM4 P18106 A0A1B6FKQ6 B3P1Q9
PDB
4E93
E-value=2.02351e-51,
Score=514
Ontologies
KEGG
GO
PANTHER
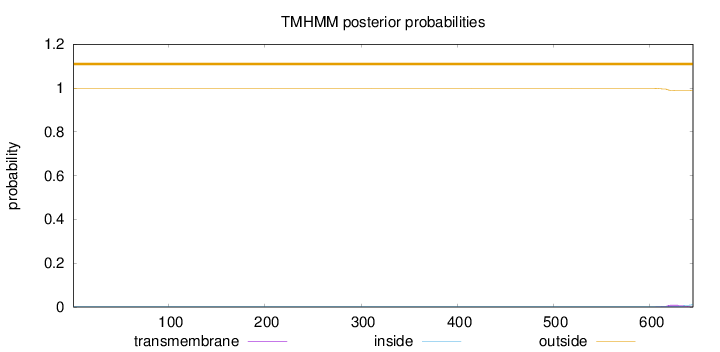
Topology
Subcellular location
Membrane
At stages 11-12 of embryogenesis, localizes to adherens junctions in epidermal cells. At stages 13-14, is no longer detected in the epidermal cell junctions which border with the amnioserosa. With evidence from 6 publications.
Cell junction At stages 11-12 of embryogenesis, localizes to adherens junctions in epidermal cells. At stages 13-14, is no longer detected in the epidermal cell junctions which border with the amnioserosa. With evidence from 6 publications.
Adherens junction At stages 11-12 of embryogenesis, localizes to adherens junctions in epidermal cells. At stages 13-14, is no longer detected in the epidermal cell junctions which border with the amnioserosa. With evidence from 6 publications.
Cell junction At stages 11-12 of embryogenesis, localizes to adherens junctions in epidermal cells. At stages 13-14, is no longer detected in the epidermal cell junctions which border with the amnioserosa. With evidence from 6 publications.
Adherens junction At stages 11-12 of embryogenesis, localizes to adherens junctions in epidermal cells. At stages 13-14, is no longer detected in the epidermal cell junctions which border with the amnioserosa. With evidence from 6 publications.
Length:
645
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.20314
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00104
outside
1 - 645
Population Genetic Test Statistics
Pi
172.741033
Theta
172.21988
Tajima's D
0.058667
CLR
0.572148
CSRT
0.385280735963202
Interpretation
Uncertain
