Gene
KWMTBOMO03303
Pre Gene Modal
BGIBMGA006423
Annotation
scolexin_precursor_[Bombyx_mori]
Full name
Scolexin B
+ More
Trypsin 3A1
Trypsin 3A1
Location in the cell
Cytoplasmic Reliability : 1.429 Extracellular Reliability : 1.126
Sequence
CDS
ATGTTCGCCAACAAACAGTCGGTTCTCGCGGTGATCGCGCTGGCGGCAAGTGCGTGCGCGCTACCTCAACCCGGCGCCAATGACATACCGGATTTAAAACAAAAATCTGCGCTGGTAACAACTGAAATTACAAAGACACAGTCTGATGTGAAAGCAGTACACGAAAGGTTTCCACATGCCGTTCTTTTCGGTGGTACCTGTGGAGGTTCCATTATTAGCCCGAAATGGATCCTCACTGCGGGACACTGCACACTGTTCACGAACGGTCATTACGTATTGGCCGGTACTAACAAATCAGACGACCAAAGCGGCATTATTCGCTACGTGAAACGAATGGTCATCCACCCGCTCTTCTCTGTTGGACCATACTGGCTTGACGTAGAAGATTTTAATCTTAAACAGGTAGCAGCTAGGTGGGACTTTTTACTGGTCGAATTGGAGGAGCCCTTACCCGTGGACGGCAAAACGATAAAAGTTGCAACACTCGATGATCAGCCAAATCTACCAATCGGAGTCGACGTCGGATATGCTGGCTACGGAACTGACGAGCATGGGGGCGTGATGCGTAAAGATATGCACGCCATGGAGCTATCAACGCAGTCCGACGAAGTATGTTCTAAATTAGAGCAATACAACTCTCTGGATATGATCTGCGCTAAAGGTCGCCCCCCACGGTTCGACTCTGCTTGCAATGGTGATAGTGGCAGCGGGCTCGTGGACGGCGAGGGGCGGCTGGTGGGGGTGGCGTCGTGGGTGGAAAATGACGCTTTTGAGTGCCGCAACGGAAACCTTGTCGTCTTCTCCAGGGTCTCCAGAGCGAGGGACTGGATCCGAGAGGTCACCGAGATTTAA
Protein
MFANKQSVLAVIALAASACALPQPGANDIPDLKQKSALVTTEITKTQSDVKAVHERFPHAVLFGGTCGGSIISPKWILTAGHCTLFTNGHYVLAGTNKSDDQSGIIRYVKRMVIHPLFSVGPYWLDVEDFNLKQVAARWDFLLVELEEPLPVDGKTIKVATLDDQPNLPIGVDVGYAGYGTDEHGGVMRKDMHAMELSTQSDEVCSKLEQYNSLDMICAKGRPPRFDSACNGDSGSGLVDGEGRLVGVASWVENDAFECRNGNLVVFSRVSRARDWIREVTEI
Summary
Description
Major function may be to aid in digestion of the blood meal.
Catalytic Activity
Preferential cleavage: Arg-|-Xaa, Lys-|-Xaa.
Similarity
Belongs to the peptidase S1 family.
Keywords
Direct protein sequencing
Disulfide bond
Hydrolase
Protease
Serine protease
Signal
Alternative splicing
Complete proteome
Digestion
Reference proteome
Secreted
Zymogen
Feature
chain Scolexin B
propeptide Activation peptide
splice variant In isoform A.
propeptide Activation peptide
splice variant In isoform A.
Uniprot
Q1HPY5
O96991
A0A2W1BRL8
C1JE15
A0A0L7L8X2
A0A1E1W1P6
+ More
O96990 A0A194Q319 A0A3G1T190 A0A212F307 S4PB00 A0A0N0PC69 A0A225VJS1 A0A0K8VJE5 A0A1B0FQA8 A0A0K8U381 D3TKK0 A0A1A9ZCT6 A0A034WPU6 A0A1A9XJL9 A0A1W4X1D3 B4KK94 Q16ID3 A0A1B0BCW7 A0A3B0JB03 A0A1A9WKF8 A0A182HBG3 B4I2Z7 B4Q9L4 A0A329SI46 A0A0A1XQQ7 P29786-2 E2BS71 Q8IQ10 P29786 A0A1B0DA37 A0A3G5BIL4 B3N333 A0A1I8NEJ2 A0A1W4V706 A0A182G5M7 A0A182UM13 V9F078 W2WWN2 W2Q3Q9 W2Z440 A0A081A1S3 W2N5V5 B4NXX8 Q0IF85 A0A1A9VW11 A0A182KQG4 A0A0W8C2U1 W2L088 Q7QIZ2 M3YX35 G9D4L8 W8C3D6 Q171L4 A0A0K8VH40 A0A225UX55 A0A1S4FH94 K3WIR9 A0A2Y9KEI8 A0A182G8J0 H3D9R1 Q4RZ78 A0A3B3ZBI4 A0A0L0CEM4 A0A315VEJ6 Q16ID4 H3H1Y5 B4KK93 D0V550 A0A182H3A3 A0A1A9XTE4 A0A329T0I7 A0A0M4EPN7 B3MP30 A0A225VKG3 Q29MC6 A0A1S4FZU0 A0A1V1FIS6 A0A182GUL7 C5IBF7 M3X005 A2TGR7 A0A212F633 M4VEY6 E5RVK3 A0A1B3TNV7 W8C5I2 A0A2C9GRR7 Q16KW4
O96990 A0A194Q319 A0A3G1T190 A0A212F307 S4PB00 A0A0N0PC69 A0A225VJS1 A0A0K8VJE5 A0A1B0FQA8 A0A0K8U381 D3TKK0 A0A1A9ZCT6 A0A034WPU6 A0A1A9XJL9 A0A1W4X1D3 B4KK94 Q16ID3 A0A1B0BCW7 A0A3B0JB03 A0A1A9WKF8 A0A182HBG3 B4I2Z7 B4Q9L4 A0A329SI46 A0A0A1XQQ7 P29786-2 E2BS71 Q8IQ10 P29786 A0A1B0DA37 A0A3G5BIL4 B3N333 A0A1I8NEJ2 A0A1W4V706 A0A182G5M7 A0A182UM13 V9F078 W2WWN2 W2Q3Q9 W2Z440 A0A081A1S3 W2N5V5 B4NXX8 Q0IF85 A0A1A9VW11 A0A182KQG4 A0A0W8C2U1 W2L088 Q7QIZ2 M3YX35 G9D4L8 W8C3D6 Q171L4 A0A0K8VH40 A0A225UX55 A0A1S4FH94 K3WIR9 A0A2Y9KEI8 A0A182G8J0 H3D9R1 Q4RZ78 A0A3B3ZBI4 A0A0L0CEM4 A0A315VEJ6 Q16ID4 H3H1Y5 B4KK93 D0V550 A0A182H3A3 A0A1A9XTE4 A0A329T0I7 A0A0M4EPN7 B3MP30 A0A225VKG3 Q29MC6 A0A1S4FZU0 A0A1V1FIS6 A0A182GUL7 C5IBF7 M3X005 A2TGR7 A0A212F633 M4VEY6 E5RVK3 A0A1B3TNV7 W8C5I2 A0A2C9GRR7 Q16KW4
EC Number
3.4.21.-
3.4.21.4
3.4.21.4
Pubmed
19121390
10210202
28756777
19442669
26227816
26354079
+ More
22118469 23622113 20353571 25348373 17994087 17510324 26483478 22936249 25830018 9087545 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30400621 25315136 17550304 20966253 12364791 14747013 17210077 21967427 24495485 15496914 25463417 26108605 29703783 16946064 20017753 15632085 28410430 19204376 17975172 23190728 27553646
22118469 23622113 20353571 25348373 17994087 17510324 26483478 22936249 25830018 9087545 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30400621 25315136 17550304 20966253 12364791 14747013 17210077 21967427 24495485 15496914 25463417 26108605 29703783 16946064 20017753 15632085 28410430 19204376 17975172 23190728 27553646
EMBL
BABH01008993
DQ443267
ABF51356.1
AF087005
AAD14592.1
KZ149939
+ More
PZC76961.1 FJ561484 ACO55200.1 JTDY01002173 KOB71952.1 GDQN01010192 JAT80862.1 AF087004 AAD14591.1 KQ459579 KPI99399.1 MG846904 AXY94756.1 AGBW02010641 OWR48128.1 GAIX01004796 JAA87764.1 KQ460650 KPJ13067.1 NBNE01004358 OWZ05585.1 GDHF01013298 JAI39016.1 CCAG010020679 GDHF01031554 JAI20760.1 EZ421932 EZ423329 ADD18228.1 ADD19605.1 GAKP01003189 JAC55763.1 CH933807 EDW12625.1 CH478086 EAT34033.1 JXJN01012155 OUUW01000004 SPP79185.1 JXUM01032511 KQ560958 KXJ80213.1 CH480820 EDW54142.1 CM000361 CM002910 EDX03625.1 KMY87907.1 MJFZ01000145 RAW36231.1 GBXI01001399 JAD12893.1 AF487426 AF508783 X64362 CH477469 GL450151 EFN81463.1 AE014134 BT023549 AAN10373.1 AAY84949.1 AJVK01028773 MK075220 AYV99623.1 CH954177 EDV57632.1 JXUM01043731 KQ561373 KXJ78808.1 ANIZ01001910 ETI44158.1 ANIX01002191 ETP14009.1 KI669591 ETN07813.1 ANIY01002292 ETP42058.1 ANJA01002024 ETO72834.1 KI686868 KI673545 KI693489 ETK84194.1 ETL37631.1 ETM44072.1 CM000157 EDW87549.1 CH477389 EAT42001.1 LNFO01005340 KUF78372.1 KI680266 ETL90777.1 AAAB01008807 EAA04161.3 AEYP01008298 JF777271 AEU11616.1 GAMC01002772 JAC03784.1 CH477451 EAT40684.1 GDHF01014131 JAI38183.1 NBNE01010492 OWY97407.1 GL376564 JXUM01047815 KQ561534 KXJ78237.1 CAAE01014957 CAG06304.1 JRES01000608 KNC29914.1 NHOQ01001935 PWA20604.1 EAT34032.1 DS566107 EDW12624.1 GU018015 ACY24348.1 JXUM01107033 KQ565105 KXJ71330.1 MJFZ01000031 RAW41402.1 CP012523 ALC38837.1 CH902620 EDV31196.1 OWZ05584.1 CH379060 EAL33768.1 FX985298 BAX07311.1 JXUM01089555 KQ563779 KXJ73325.1 FJ839347 ACR61328.1 AANG04003369 EF199627 ABM88498.1 AGBW02010081 OWR49201.1 CP003537 AGH96601.1 AB604648 BAJ46146.1 KU925715 AOG75340.1 GAMC01001732 JAC04824.1 APCN01000169 CH477936 EAT34942.1
PZC76961.1 FJ561484 ACO55200.1 JTDY01002173 KOB71952.1 GDQN01010192 JAT80862.1 AF087004 AAD14591.1 KQ459579 KPI99399.1 MG846904 AXY94756.1 AGBW02010641 OWR48128.1 GAIX01004796 JAA87764.1 KQ460650 KPJ13067.1 NBNE01004358 OWZ05585.1 GDHF01013298 JAI39016.1 CCAG010020679 GDHF01031554 JAI20760.1 EZ421932 EZ423329 ADD18228.1 ADD19605.1 GAKP01003189 JAC55763.1 CH933807 EDW12625.1 CH478086 EAT34033.1 JXJN01012155 OUUW01000004 SPP79185.1 JXUM01032511 KQ560958 KXJ80213.1 CH480820 EDW54142.1 CM000361 CM002910 EDX03625.1 KMY87907.1 MJFZ01000145 RAW36231.1 GBXI01001399 JAD12893.1 AF487426 AF508783 X64362 CH477469 GL450151 EFN81463.1 AE014134 BT023549 AAN10373.1 AAY84949.1 AJVK01028773 MK075220 AYV99623.1 CH954177 EDV57632.1 JXUM01043731 KQ561373 KXJ78808.1 ANIZ01001910 ETI44158.1 ANIX01002191 ETP14009.1 KI669591 ETN07813.1 ANIY01002292 ETP42058.1 ANJA01002024 ETO72834.1 KI686868 KI673545 KI693489 ETK84194.1 ETL37631.1 ETM44072.1 CM000157 EDW87549.1 CH477389 EAT42001.1 LNFO01005340 KUF78372.1 KI680266 ETL90777.1 AAAB01008807 EAA04161.3 AEYP01008298 JF777271 AEU11616.1 GAMC01002772 JAC03784.1 CH477451 EAT40684.1 GDHF01014131 JAI38183.1 NBNE01010492 OWY97407.1 GL376564 JXUM01047815 KQ561534 KXJ78237.1 CAAE01014957 CAG06304.1 JRES01000608 KNC29914.1 NHOQ01001935 PWA20604.1 EAT34032.1 DS566107 EDW12624.1 GU018015 ACY24348.1 JXUM01107033 KQ565105 KXJ71330.1 MJFZ01000031 RAW41402.1 CP012523 ALC38837.1 CH902620 EDV31196.1 OWZ05584.1 CH379060 EAL33768.1 FX985298 BAX07311.1 JXUM01089555 KQ563779 KXJ73325.1 FJ839347 ACR61328.1 AANG04003369 EF199627 ABM88498.1 AGBW02010081 OWR49201.1 CP003537 AGH96601.1 AB604648 BAJ46146.1 KU925715 AOG75340.1 GAMC01001732 JAC04824.1 APCN01000169 CH477936 EAT34942.1
Proteomes
UP000005204
UP000037510
UP000053268
UP000007151
UP000053240
UP000198211
+ More
UP000092444 UP000092445 UP000092443 UP000192223 UP000009192 UP000008820 UP000092460 UP000268350 UP000091820 UP000069940 UP000249989 UP000001292 UP000000304 UP000251314 UP000008237 UP000000803 UP000092462 UP000008711 UP000095301 UP000192221 UP000075903 UP000018721 UP000018958 UP000018817 UP000018948 UP000028582 UP000053236 UP000053864 UP000054532 UP000002282 UP000078200 UP000075882 UP000052943 UP000054423 UP000007062 UP000000715 UP000248482 UP000007303 UP000261520 UP000037069 UP000005238 UP000092553 UP000007801 UP000001819 UP000011712 UP000075840
UP000092444 UP000092445 UP000092443 UP000192223 UP000009192 UP000008820 UP000092460 UP000268350 UP000091820 UP000069940 UP000249989 UP000001292 UP000000304 UP000251314 UP000008237 UP000000803 UP000092462 UP000008711 UP000095301 UP000192221 UP000075903 UP000018721 UP000018958 UP000018817 UP000018948 UP000028582 UP000053236 UP000053864 UP000054532 UP000002282 UP000078200 UP000075882 UP000052943 UP000054423 UP000007062 UP000000715 UP000248482 UP000007303 UP000261520 UP000037069 UP000005238 UP000092553 UP000007801 UP000001819 UP000011712 UP000075840
Interpro
Gene 3D
ProteinModelPortal
Q1HPY5
O96991
A0A2W1BRL8
C1JE15
A0A0L7L8X2
A0A1E1W1P6
+ More
O96990 A0A194Q319 A0A3G1T190 A0A212F307 S4PB00 A0A0N0PC69 A0A225VJS1 A0A0K8VJE5 A0A1B0FQA8 A0A0K8U381 D3TKK0 A0A1A9ZCT6 A0A034WPU6 A0A1A9XJL9 A0A1W4X1D3 B4KK94 Q16ID3 A0A1B0BCW7 A0A3B0JB03 A0A1A9WKF8 A0A182HBG3 B4I2Z7 B4Q9L4 A0A329SI46 A0A0A1XQQ7 P29786-2 E2BS71 Q8IQ10 P29786 A0A1B0DA37 A0A3G5BIL4 B3N333 A0A1I8NEJ2 A0A1W4V706 A0A182G5M7 A0A182UM13 V9F078 W2WWN2 W2Q3Q9 W2Z440 A0A081A1S3 W2N5V5 B4NXX8 Q0IF85 A0A1A9VW11 A0A182KQG4 A0A0W8C2U1 W2L088 Q7QIZ2 M3YX35 G9D4L8 W8C3D6 Q171L4 A0A0K8VH40 A0A225UX55 A0A1S4FH94 K3WIR9 A0A2Y9KEI8 A0A182G8J0 H3D9R1 Q4RZ78 A0A3B3ZBI4 A0A0L0CEM4 A0A315VEJ6 Q16ID4 H3H1Y5 B4KK93 D0V550 A0A182H3A3 A0A1A9XTE4 A0A329T0I7 A0A0M4EPN7 B3MP30 A0A225VKG3 Q29MC6 A0A1S4FZU0 A0A1V1FIS6 A0A182GUL7 C5IBF7 M3X005 A2TGR7 A0A212F633 M4VEY6 E5RVK3 A0A1B3TNV7 W8C5I2 A0A2C9GRR7 Q16KW4
O96990 A0A194Q319 A0A3G1T190 A0A212F307 S4PB00 A0A0N0PC69 A0A225VJS1 A0A0K8VJE5 A0A1B0FQA8 A0A0K8U381 D3TKK0 A0A1A9ZCT6 A0A034WPU6 A0A1A9XJL9 A0A1W4X1D3 B4KK94 Q16ID3 A0A1B0BCW7 A0A3B0JB03 A0A1A9WKF8 A0A182HBG3 B4I2Z7 B4Q9L4 A0A329SI46 A0A0A1XQQ7 P29786-2 E2BS71 Q8IQ10 P29786 A0A1B0DA37 A0A3G5BIL4 B3N333 A0A1I8NEJ2 A0A1W4V706 A0A182G5M7 A0A182UM13 V9F078 W2WWN2 W2Q3Q9 W2Z440 A0A081A1S3 W2N5V5 B4NXX8 Q0IF85 A0A1A9VW11 A0A182KQG4 A0A0W8C2U1 W2L088 Q7QIZ2 M3YX35 G9D4L8 W8C3D6 Q171L4 A0A0K8VH40 A0A225UX55 A0A1S4FH94 K3WIR9 A0A2Y9KEI8 A0A182G8J0 H3D9R1 Q4RZ78 A0A3B3ZBI4 A0A0L0CEM4 A0A315VEJ6 Q16ID4 H3H1Y5 B4KK93 D0V550 A0A182H3A3 A0A1A9XTE4 A0A329T0I7 A0A0M4EPN7 B3MP30 A0A225VKG3 Q29MC6 A0A1S4FZU0 A0A1V1FIS6 A0A182GUL7 C5IBF7 M3X005 A2TGR7 A0A212F633 M4VEY6 E5RVK3 A0A1B3TNV7 W8C5I2 A0A2C9GRR7 Q16KW4
PDB
3SO3
E-value=2.35355e-11,
Score=164
Ontologies
GO
Topology
Subcellular location
Secreted
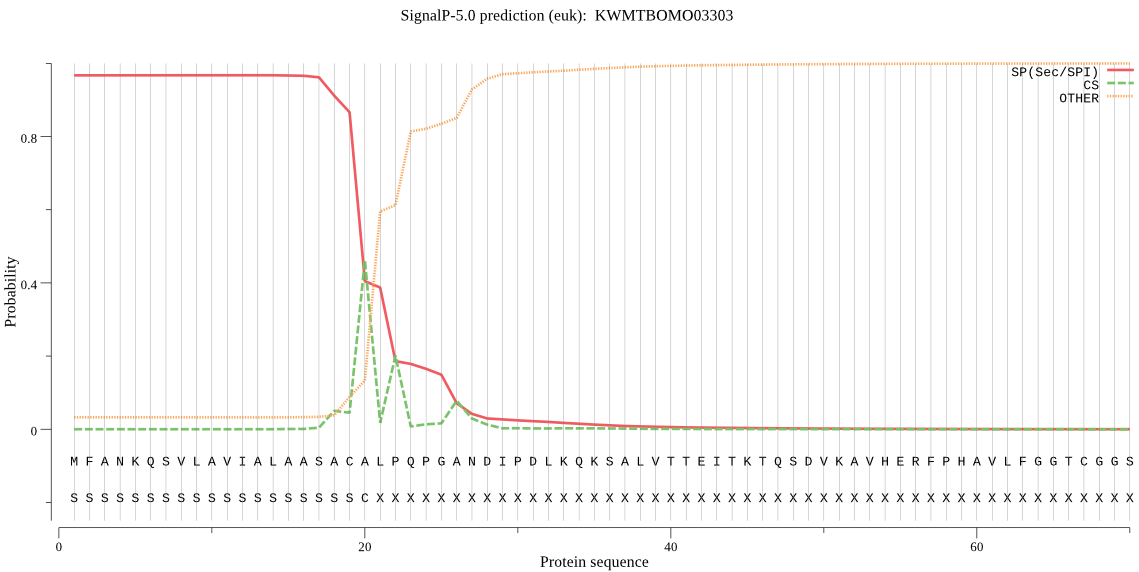
SignalP
Position: 1 - 20,
Likelihood: 0.967408
Length:
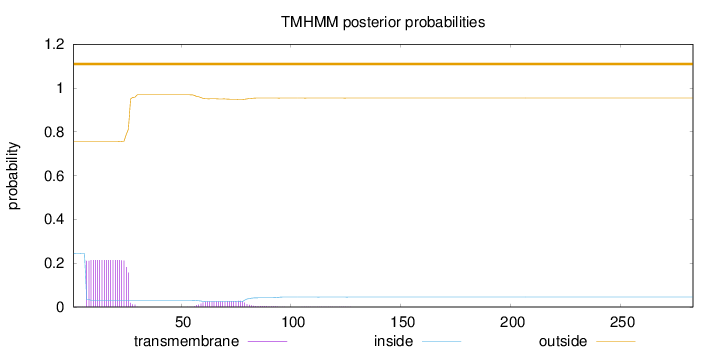
283
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.87521
Exp number, first 60 AAs:
4.29629
Total prob of N-in:
0.24508
outside
1 - 283
Population Genetic Test Statistics
Pi
197.017135
Theta
169.170455
Tajima's D
-0.900366
CLR
1.199575
CSRT
0.156442177891105
Interpretation
Uncertain
