Gene
KWMTBOMO03301 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006424
Annotation
phenylalanyl-tRNA_synthetase_beta_subunit_[Bombyx_mori]
Full name
Phenylalanine--tRNA ligase beta subunit
Alternative Name
Phenylalanyl-tRNA synthetase beta subunit
Location in the cell
Cytoplasmic Reliability : 2.538
Sequence
CDS
ATGCCCACAATTTCTTTAAAGCGCGATGCGCTTTTCGCCGCACTCGGTTCTACATACACTGATGAAGAATTTCAAGACCTATGTTTCAAATTTGGTTTAGAGCTTGATGAAGTTACTACGGAGAAACAAATGTTAATTAAAGAACAAGGTGACCAAGCAGATGCTGAATTATCTGATGAGATCCTCTACAGAATAGACATCCCAGCCAATAGATATGACTTATTGTGCCTCGAAGGACTTGTTGATGGTCTATTAGTGTTCCAGGGGAAGAAACCTCCACCTCAGTACAAAGTGAAAAAATATGAAGACTGTAACTCTTTACACCTAACGATGGCCACAGCACAAATAAGACCCTATGCTGTAGCAGCTGTACTTAGAGGCATTACTTTTACAAAGGAATCTTATGATAGTTTCATTGATCTACAGGACAAACTGCACCAAAACATTTGCAGGAAAAGGACTTTAGTGGCTATAGGAACACATGATCTGGACACTATACATGGTCCATTCATCTATGATGCATTGCCTCCAAATGAGATTAAGTTCAAAGCTCTTAATCAGCCAAAAGAATTAACTGCACCTGAATTAATGGAGTTATATTCGAATCACGCCCAATTAAAACAATATTTAGGAATCATAAAGGAGAGCCCAGTGTATCCAATCATCAAAGATAAAAACGGAGTGATATTGTCTATGCCTCCAATAATTAATAGTGATCATTCTAAAATTACACTAAATACAAAGAATGTATTCATAGAATGCACTGCAACTGATTTAACTAAGGCTATTGTAGTTCTGGACACGGTGGTCAGCATGTTCTCGAAATACTGCACGAGCGAGTATGAAGTACAACAATGTAAAGTTTTCTCTCCTGATGGAACTTACGAATTGTATCCCAAGCTTCAATATAGAGAAGAACTTATTAATGTTGATAAAGCCAACAACTATATTGGGATCAGTGAAGAAGGTGATAAGCTAGCGTCGTTGCTGTCCCGGATGTGTCTCCAGACCGCGCACGAGGGCTCCGTGCTGCGCGTGCGAGTCCCCCCCACGAGGCACGACGTCATTCACGCTTGCGACCTCTACGAAGACATCGCTATAGCATACGGGTACAACCGCATAGCGCGGCGCCCCGCCCGCGCCGTGACGTCAGGCGGACAGGACCCCGCCAACAAACTCACGGAGCAGCTGCGGAACGAGTGCGCCCGCGCCGGCTACACTGAAGCGCTCACCTTCACACTTTGCTCCCGTGAAGACGTATCTACGAAATTGGGTGTTAAAATCGAAGATGTACCGGCCGCCCACATCTCCAATCCCAAGACCCTGGAGTTCCAAGTAGTCCGAACGTTGTTACTACCCGGGCTGCTGAAAACCATTGCTGCAAACAAAAAAATGCCTTTACCCTTGAAACTCTTCGAGATCAGCGATGTTGTGCTTCTAGATGAGAATGCCGAAAGCGGGGCACGCAACGTGCGGCGCGCGTGCGGCGTGCACTGCGGGCGCGCGGCCGGCTTCCAGCACGTGCACGGCCTGCTCGACCGGCTCATGGCGCAGCTGCGGGTGCGGCACCGCGACCAGTACGCGCTGCGACCTGCGCAAGATCCCGCTTATTTCCCTGGTCGCTGCGCTGAAGTTGTACTTCAAGGAAAAGTAATCGGAAAGATTGGAGTCATCCACCCGAATGTGCTGACAGCATTCGAGCTTACGAATCCGTGCTCTGCTGTTGAAATAGATATTGAACCATTCGTGTAA
Protein
MPTISLKRDALFAALGSTYTDEEFQDLCFKFGLELDEVTTEKQMLIKEQGDQADAELSDEILYRIDIPANRYDLLCLEGLVDGLLVFQGKKPPPQYKVKKYEDCNSLHLTMATAQIRPYAVAAVLRGITFTKESYDSFIDLQDKLHQNICRKRTLVAIGTHDLDTIHGPFIYDALPPNEIKFKALNQPKELTAPELMELYSNHAQLKQYLGIIKESPVYPIIKDKNGVILSMPPIINSDHSKITLNTKNVFIECTATDLTKAIVVLDTVVSMFSKYCTSEYEVQQCKVFSPDGTYELYPKLQYREELINVDKANNYIGISEEGDKLASLLSRMCLQTAHEGSVLRVRVPPTRHDVIHACDLYEDIAIAYGYNRIARRPARAVTSGGQDPANKLTEQLRNECARAGYTEALTFTLCSREDVSTKLGVKIEDVPAAHISNPKTLEFQVVRTLLLPGLLKTIAANKKMPLPLKLFEISDVVLLDENAESGARNVRRACGVHCGRAAGFQHVHGLLDRLMAQLRVRHRDQYALRPAQDPAYFPGRCAEVVLQGKVIGKIGVIHPNVLTAFELTNPCSAVEIDIEPFV
Summary
Catalytic Activity
ATP + L-phenylalanine + tRNA(Phe) = AMP + diphosphate + H(+) + L-phenylalanyl-tRNA(Phe)
Subunit
Tetramer of two alpha and two beta subunits.
Similarity
Belongs to the GST superfamily.
Belongs to the phenylalanyl-tRNA synthetase beta subunit family. Type 2 subfamily.
Belongs to the EF-1-beta/EF-1-delta family.
Belongs to the phenylalanyl-tRNA synthetase beta subunit family. Type 2 subfamily.
Belongs to the EF-1-beta/EF-1-delta family.
Keywords
Aminoacyl-tRNA synthetase
ATP-binding
Complete proteome
Cytoplasm
Ligase
Nucleotide-binding
Protein biosynthesis
Reference proteome
Feature
chain Phenylalanine--tRNA ligase beta subunit
Uniprot
E0D4V7
A0A2W1BWD3
A0A2H1X2X4
A0A212F320
S4PK77
A0A0N1PGH5
+ More
Q9VCA5 B4HGK9 A0A0T6B0S1 A0A1Y1L114 B4PKZ2 A0A2J7PZJ6 B3P768 A0A1W4US24 B3M0J6 A0A2P8XNY2 B4M5Q8 A0A2J7PZJ4 B4NAF8 B4K7G4 U5EXX1 Q298I1 A0A139WN72 A0A1W4XEQ4 A0A3B0K3Y3 B4G3Z1 B4QT31 A0A0L0CRD1 A0A084W1R2 A0A182JMM9 W8BG13 A0A1B6M1Z7 A0A0A1XCZ9 A0A182NSK1 A0A1L8DFL7 A0A1B6EWG0 Q7QAV9 A0A182HZ48 A0A0K8VJ14 A0A182URB8 A0A182KXB4 A0A034W5K7 A0A182JYD1 A0A182WVZ0 A0A1I8MRV2 B4JH33 A0A182QFA5 A0A182P097 A0A182TSZ9 A0A1B6DE20 T1PJJ9 W5J571 A0A1I8Q7C3 A0A023EU76 A0A182GPY4 A0A182R8A1 A0A2M4A5N1 A0A2M4A5P1 A0A182VS39 A0A2M4BI26 A0A1B0CSF9 A0A2M4BIC9 A0A2M3YZU6 A0A182FL19 A0A026WAP6 A0A2M3Z001 A0A2M4A5W5 Q17C95 A0A1S4F853 A0A1Q3F447 A0A182YLK9 N6U1A5 A0A023EVD9 A0A182M295 J9K9N3 E2BE89 A0A1J1HYI0 A0A2S2R343 A0A3B4ZPN7 E2A2Q0 A0A1A9XG67 A0A1B0BQ40 A0A1A9WY64 A0A2M4BEX5 A0A232EZT4 A0A3P8RQ16 E9GZV7 A0A2J7PZK5 H2LYY4 A0A3P9IPY4 A0A1A9UZ08 A0A3B3HKC4 H2LZ90 Q5EBG3 A0A0L7RD93 A0A3B4U448 A0A3P9GY17 A0A3B4ZCP1 F7DMT3 Q7T0M8
Q9VCA5 B4HGK9 A0A0T6B0S1 A0A1Y1L114 B4PKZ2 A0A2J7PZJ6 B3P768 A0A1W4US24 B3M0J6 A0A2P8XNY2 B4M5Q8 A0A2J7PZJ4 B4NAF8 B4K7G4 U5EXX1 Q298I1 A0A139WN72 A0A1W4XEQ4 A0A3B0K3Y3 B4G3Z1 B4QT31 A0A0L0CRD1 A0A084W1R2 A0A182JMM9 W8BG13 A0A1B6M1Z7 A0A0A1XCZ9 A0A182NSK1 A0A1L8DFL7 A0A1B6EWG0 Q7QAV9 A0A182HZ48 A0A0K8VJ14 A0A182URB8 A0A182KXB4 A0A034W5K7 A0A182JYD1 A0A182WVZ0 A0A1I8MRV2 B4JH33 A0A182QFA5 A0A182P097 A0A182TSZ9 A0A1B6DE20 T1PJJ9 W5J571 A0A1I8Q7C3 A0A023EU76 A0A182GPY4 A0A182R8A1 A0A2M4A5N1 A0A2M4A5P1 A0A182VS39 A0A2M4BI26 A0A1B0CSF9 A0A2M4BIC9 A0A2M3YZU6 A0A182FL19 A0A026WAP6 A0A2M3Z001 A0A2M4A5W5 Q17C95 A0A1S4F853 A0A1Q3F447 A0A182YLK9 N6U1A5 A0A023EVD9 A0A182M295 J9K9N3 E2BE89 A0A1J1HYI0 A0A2S2R343 A0A3B4ZPN7 E2A2Q0 A0A1A9XG67 A0A1B0BQ40 A0A1A9WY64 A0A2M4BEX5 A0A232EZT4 A0A3P8RQ16 E9GZV7 A0A2J7PZK5 H2LYY4 A0A3P9IPY4 A0A1A9UZ08 A0A3B3HKC4 H2LZ90 Q5EBG3 A0A0L7RD93 A0A3B4U448 A0A3P9GY17 A0A3B4ZCP1 F7DMT3 Q7T0M8
EC Number
6.1.1.20
Pubmed
19121390
28756777
22118469
23622113
26354079
10731132
+ More
12537572 12537569 25427601 17994087 28004739 17550304 29403074 15632085 18362917 19820115 26108605 24438588 24495485 25830018 12364791 14747013 17210077 20966253 25348373 25315136 20920257 23761445 24945155 26483478 24508170 17510324 25244985 23537049 20798317 28648823 21292972 17554307 20431018
12537572 12537569 25427601 17994087 28004739 17550304 29403074 15632085 18362917 19820115 26108605 24438588 24495485 25830018 12364791 14747013 17210077 20966253 25348373 25315136 20920257 23761445 24945155 26483478 24508170 17510324 25244985 23537049 20798317 28648823 21292972 17554307 20431018
EMBL
BABH01008989
BABH01008990
BABH01008991
AB546862
BAJ14787.1
KZ149939
+ More
PZC76976.1 ODYU01013030 SOQ59665.1 AGBW02010641 OWR48124.1 GAIX01004655 JAA87905.1 KQ460650 KPJ13070.1 AE014297 AY052086 CH480815 EDW43462.1 LJIG01016334 KRT80914.1 GEZM01067984 JAV67284.1 CM000160 EDW98977.1 NEVH01020334 PNF21755.1 CH954182 EDV53888.1 CH902617 EDV44243.1 PYGN01001634 PSN33654.1 CH940652 EDW58984.1 PNF21751.1 CH964232 EDW80772.1 CH933806 EDW14288.1 GANO01000702 JAB59169.1 CM000070 EAL27974.1 KQ971312 KYB29382.1 OUUW01000007 SPP82680.1 CH479179 EDW24402.1 CM000364 EDX14195.1 JRES01000007 KNC34920.1 ATLV01019415 KE525269 KFB44156.1 GAMC01006320 JAC00236.1 GEBQ01010008 JAT29969.1 GBXI01005949 JAD08343.1 GFDF01008848 JAV05236.1 GECZ01027521 JAS42248.1 AAAB01008888 EAA08937.2 EGK97049.1 APCN01005482 GDHF01013764 JAI38550.1 GAKP01008096 JAC50856.1 CH916369 EDV92724.1 AXCN02000234 GEDC01013416 JAS23882.1 KA648108 AFP62737.1 ADMH02002130 ETN58558.1 GAPW01000643 JAC12955.1 JXUM01015409 KQ560429 KXJ82448.1 GGFK01002701 MBW36022.1 GGFK01002711 MBW36032.1 GGFJ01003569 MBW52710.1 AJWK01026035 GGFJ01003570 MBW52711.1 GGFM01001048 MBW21799.1 KK107295 EZA53110.1 GGFM01001075 MBW21826.1 GGFK01002799 MBW36120.1 CH477310 EAT43980.1 GFDL01012720 JAV22325.1 APGK01043586 KB741015 ENN75330.1 GAPW01000641 JAC12957.1 AXCM01000846 ABLF02014205 ABLF02014206 GL447768 EFN85934.1 CVRI01000026 CRK92388.1 GGMS01015182 MBY84385.1 GL436195 EFN72288.1 JXJN01018373 GGFJ01002453 MBW51594.1 NNAY01001458 OXU23902.1 GL732579 EFX74882.1 PNF21752.1 BC089642 CR942340 AAH89642.1 CAJ83855.1 KQ414613 KOC68937.1 AAMC01060088 AAMC01060089 AAMC01060090 AAMC01060091 AAMC01060092 BC056121 AAH56121.1
PZC76976.1 ODYU01013030 SOQ59665.1 AGBW02010641 OWR48124.1 GAIX01004655 JAA87905.1 KQ460650 KPJ13070.1 AE014297 AY052086 CH480815 EDW43462.1 LJIG01016334 KRT80914.1 GEZM01067984 JAV67284.1 CM000160 EDW98977.1 NEVH01020334 PNF21755.1 CH954182 EDV53888.1 CH902617 EDV44243.1 PYGN01001634 PSN33654.1 CH940652 EDW58984.1 PNF21751.1 CH964232 EDW80772.1 CH933806 EDW14288.1 GANO01000702 JAB59169.1 CM000070 EAL27974.1 KQ971312 KYB29382.1 OUUW01000007 SPP82680.1 CH479179 EDW24402.1 CM000364 EDX14195.1 JRES01000007 KNC34920.1 ATLV01019415 KE525269 KFB44156.1 GAMC01006320 JAC00236.1 GEBQ01010008 JAT29969.1 GBXI01005949 JAD08343.1 GFDF01008848 JAV05236.1 GECZ01027521 JAS42248.1 AAAB01008888 EAA08937.2 EGK97049.1 APCN01005482 GDHF01013764 JAI38550.1 GAKP01008096 JAC50856.1 CH916369 EDV92724.1 AXCN02000234 GEDC01013416 JAS23882.1 KA648108 AFP62737.1 ADMH02002130 ETN58558.1 GAPW01000643 JAC12955.1 JXUM01015409 KQ560429 KXJ82448.1 GGFK01002701 MBW36022.1 GGFK01002711 MBW36032.1 GGFJ01003569 MBW52710.1 AJWK01026035 GGFJ01003570 MBW52711.1 GGFM01001048 MBW21799.1 KK107295 EZA53110.1 GGFM01001075 MBW21826.1 GGFK01002799 MBW36120.1 CH477310 EAT43980.1 GFDL01012720 JAV22325.1 APGK01043586 KB741015 ENN75330.1 GAPW01000641 JAC12957.1 AXCM01000846 ABLF02014205 ABLF02014206 GL447768 EFN85934.1 CVRI01000026 CRK92388.1 GGMS01015182 MBY84385.1 GL436195 EFN72288.1 JXJN01018373 GGFJ01002453 MBW51594.1 NNAY01001458 OXU23902.1 GL732579 EFX74882.1 PNF21752.1 BC089642 CR942340 AAH89642.1 CAJ83855.1 KQ414613 KOC68937.1 AAMC01060088 AAMC01060089 AAMC01060090 AAMC01060091 AAMC01060092 BC056121 AAH56121.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000000803
UP000001292
UP000002282
+ More
UP000235965 UP000008711 UP000192221 UP000007801 UP000245037 UP000008792 UP000007798 UP000009192 UP000001819 UP000007266 UP000192223 UP000268350 UP000008744 UP000000304 UP000037069 UP000030765 UP000075880 UP000075884 UP000007062 UP000075840 UP000075903 UP000075882 UP000075881 UP000076407 UP000095301 UP000001070 UP000075886 UP000075885 UP000075902 UP000000673 UP000095300 UP000069940 UP000249989 UP000075900 UP000075920 UP000092461 UP000069272 UP000053097 UP000008820 UP000076408 UP000019118 UP000075883 UP000007819 UP000008237 UP000183832 UP000261400 UP000000311 UP000092443 UP000092460 UP000091820 UP000215335 UP000265080 UP000000305 UP000001038 UP000265200 UP000078200 UP000053825 UP000261420 UP000261360 UP000008143
UP000235965 UP000008711 UP000192221 UP000007801 UP000245037 UP000008792 UP000007798 UP000009192 UP000001819 UP000007266 UP000192223 UP000268350 UP000008744 UP000000304 UP000037069 UP000030765 UP000075880 UP000075884 UP000007062 UP000075840 UP000075903 UP000075882 UP000075881 UP000076407 UP000095301 UP000001070 UP000075886 UP000075885 UP000075902 UP000000673 UP000095300 UP000069940 UP000249989 UP000075900 UP000075920 UP000092461 UP000069272 UP000053097 UP000008820 UP000076408 UP000019118 UP000075883 UP000007819 UP000008237 UP000183832 UP000261400 UP000000311 UP000092443 UP000092460 UP000091820 UP000215335 UP000265080 UP000000305 UP000001038 UP000265200 UP000078200 UP000053825 UP000261420 UP000261360 UP000008143
Pfam
Interpro
IPR005147
tRNA_synthase_B5-dom
+ More
IPR004531 Phe-tRNA-synth_IIc_bsu_arc
IPR040659 PhetRS_B1
IPR005146 B3/B4_tRNA-bd
IPR041616 PheRS_beta_core
IPR009061 DNA-bd_dom_put_sf
IPR020825 Phe-tRNA_synthase_B3/B4
IPR036249 Thioredoxin-like_sf
IPR036282 Glutathione-S-Trfase_C_sf
IPR004046 GST_C
IPR010987 Glutathione-S-Trfase_C-like
IPR040079 Glutathione_S-Trfase
IPR004045 Glutathione_S-Trfase_N
IPR011425 Med9
IPR019258 Mediator_Med4
IPR018940 EF-1_beta_acid_region_euk
IPR036219 eEF-1beta-like_sf
IPR014038 EF1B_bsu/dsu_GNE
IPR001326 Transl_elong_EF1B_B/D_CS
IPR014717 Transl_elong_EF1B/ribosomal_S6
IPR004531 Phe-tRNA-synth_IIc_bsu_arc
IPR040659 PhetRS_B1
IPR005146 B3/B4_tRNA-bd
IPR041616 PheRS_beta_core
IPR009061 DNA-bd_dom_put_sf
IPR020825 Phe-tRNA_synthase_B3/B4
IPR036249 Thioredoxin-like_sf
IPR036282 Glutathione-S-Trfase_C_sf
IPR004046 GST_C
IPR010987 Glutathione-S-Trfase_C-like
IPR040079 Glutathione_S-Trfase
IPR004045 Glutathione_S-Trfase_N
IPR011425 Med9
IPR019258 Mediator_Med4
IPR018940 EF-1_beta_acid_region_euk
IPR036219 eEF-1beta-like_sf
IPR014038 EF1B_bsu/dsu_GNE
IPR001326 Transl_elong_EF1B_B/D_CS
IPR014717 Transl_elong_EF1B/ribosomal_S6
Gene 3D
ProteinModelPortal
E0D4V7
A0A2W1BWD3
A0A2H1X2X4
A0A212F320
S4PK77
A0A0N1PGH5
+ More
Q9VCA5 B4HGK9 A0A0T6B0S1 A0A1Y1L114 B4PKZ2 A0A2J7PZJ6 B3P768 A0A1W4US24 B3M0J6 A0A2P8XNY2 B4M5Q8 A0A2J7PZJ4 B4NAF8 B4K7G4 U5EXX1 Q298I1 A0A139WN72 A0A1W4XEQ4 A0A3B0K3Y3 B4G3Z1 B4QT31 A0A0L0CRD1 A0A084W1R2 A0A182JMM9 W8BG13 A0A1B6M1Z7 A0A0A1XCZ9 A0A182NSK1 A0A1L8DFL7 A0A1B6EWG0 Q7QAV9 A0A182HZ48 A0A0K8VJ14 A0A182URB8 A0A182KXB4 A0A034W5K7 A0A182JYD1 A0A182WVZ0 A0A1I8MRV2 B4JH33 A0A182QFA5 A0A182P097 A0A182TSZ9 A0A1B6DE20 T1PJJ9 W5J571 A0A1I8Q7C3 A0A023EU76 A0A182GPY4 A0A182R8A1 A0A2M4A5N1 A0A2M4A5P1 A0A182VS39 A0A2M4BI26 A0A1B0CSF9 A0A2M4BIC9 A0A2M3YZU6 A0A182FL19 A0A026WAP6 A0A2M3Z001 A0A2M4A5W5 Q17C95 A0A1S4F853 A0A1Q3F447 A0A182YLK9 N6U1A5 A0A023EVD9 A0A182M295 J9K9N3 E2BE89 A0A1J1HYI0 A0A2S2R343 A0A3B4ZPN7 E2A2Q0 A0A1A9XG67 A0A1B0BQ40 A0A1A9WY64 A0A2M4BEX5 A0A232EZT4 A0A3P8RQ16 E9GZV7 A0A2J7PZK5 H2LYY4 A0A3P9IPY4 A0A1A9UZ08 A0A3B3HKC4 H2LZ90 Q5EBG3 A0A0L7RD93 A0A3B4U448 A0A3P9GY17 A0A3B4ZCP1 F7DMT3 Q7T0M8
Q9VCA5 B4HGK9 A0A0T6B0S1 A0A1Y1L114 B4PKZ2 A0A2J7PZJ6 B3P768 A0A1W4US24 B3M0J6 A0A2P8XNY2 B4M5Q8 A0A2J7PZJ4 B4NAF8 B4K7G4 U5EXX1 Q298I1 A0A139WN72 A0A1W4XEQ4 A0A3B0K3Y3 B4G3Z1 B4QT31 A0A0L0CRD1 A0A084W1R2 A0A182JMM9 W8BG13 A0A1B6M1Z7 A0A0A1XCZ9 A0A182NSK1 A0A1L8DFL7 A0A1B6EWG0 Q7QAV9 A0A182HZ48 A0A0K8VJ14 A0A182URB8 A0A182KXB4 A0A034W5K7 A0A182JYD1 A0A182WVZ0 A0A1I8MRV2 B4JH33 A0A182QFA5 A0A182P097 A0A182TSZ9 A0A1B6DE20 T1PJJ9 W5J571 A0A1I8Q7C3 A0A023EU76 A0A182GPY4 A0A182R8A1 A0A2M4A5N1 A0A2M4A5P1 A0A182VS39 A0A2M4BI26 A0A1B0CSF9 A0A2M4BIC9 A0A2M3YZU6 A0A182FL19 A0A026WAP6 A0A2M3Z001 A0A2M4A5W5 Q17C95 A0A1S4F853 A0A1Q3F447 A0A182YLK9 N6U1A5 A0A023EVD9 A0A182M295 J9K9N3 E2BE89 A0A1J1HYI0 A0A2S2R343 A0A3B4ZPN7 E2A2Q0 A0A1A9XG67 A0A1B0BQ40 A0A1A9WY64 A0A2M4BEX5 A0A232EZT4 A0A3P8RQ16 E9GZV7 A0A2J7PZK5 H2LYY4 A0A3P9IPY4 A0A1A9UZ08 A0A3B3HKC4 H2LZ90 Q5EBG3 A0A0L7RD93 A0A3B4U448 A0A3P9GY17 A0A3B4ZCP1 F7DMT3 Q7T0M8
PDB
3L4G
E-value=0,
Score=1712
Ontologies
GO
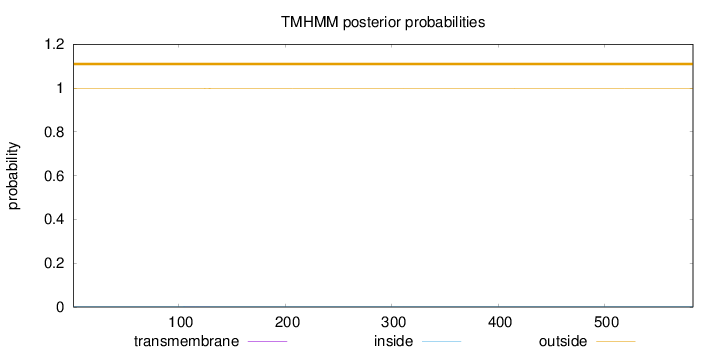
Topology
Subcellular location
Cytoplasm
Length:
583
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0216899999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00192
outside
1 - 583
Population Genetic Test Statistics
Pi
138.271601
Theta
150.932417
Tajima's D
-0.905174
CLR
1.127941
CSRT
0.157542122893855
Interpretation
Uncertain
