Gene
KWMTBOMO03274
Pre Gene Modal
BGIBMGA014467
Annotation
CCHamide_precursor_[Bombyx_mori]
Full name
Neuropeptide CCHamide-2
Location in the cell
Extracellular Reliability : 1.644 Nuclear Reliability : 1.882
Sequence
CDS
ATGGCTCAGATCTACTTAGCGGTGTCCATCGCAGCCTTGCTGATGATGTCACAAGGAGTGAGTGCTAAACGAGGCTGTTCAGCATTTGGGCACTCCTGCTTCGGCGGGCATGGAAAGAGATCAGGCGAACCTGCACCGATGGATATGGCGAATCAGGATATGATGGTGCGTCACCAGCTCGGTCAAGAGGAGACTCCTCCTCACCCAGGGTACCCTCATTCCAGCTACAACGTGCTGCAGCCTGGCGACGACATCATTCCTATTAGGGATGGAGGTGTGTACGACCATGACGCCGCTGCCAGAGACGTGATGAAGTACAAGCTTAGGAATATCTTCAAACATTGGATGGACAACTACCGACGGTCGCAGAACACAAACGACGAGTATTTCCTAGAAACGATTTAA
Protein
MAQIYLAVSIAALLMMSQGVSAKRGCSAFGHSCFGGHGKRSGEPAPMDMANQDMMVRHQLGQEETPPHPGYPHSSYNVLQPGDDIIPIRDGGVYDHDAAARDVMKYKLRNIFKHWMDNYRRSQNTNDEYFLETI
Summary
Description
Ligand for the CCHamide-2 receptor CCHa2-R.
Keywords
Amidation
Cleavage on pair of basic residues
Direct protein sequencing
Disulfide bond
Neuropeptide
Secreted
Signal
Feature
peptide Neuropeptide CCHamide-2
Uniprot
B3XYE5
A0A2W1B8P0
C0HKT1
M4QFY3
A0A385H8U2
A0A385H8U4
+ More
A0A1E1W802 A0A0L7LBS9 A0A0S1U0C3 A0A2A4J888 A0A2H1W587 A0A0C5BQ59 A0A194Q1M5 A0A0N0PCK1 A0A212ERE8 A0A1Y1LSL1 U5ELW0 B3P145 A0A1B6CDQ8 A0A1B0CG21 A0A1L8DNL3 A0A1L8D9Y1 A0A1L8DNM9 A0A1L8D9Z1 A0A1B0D9G5 A0A0L0CH44 B4PQ10 A0A1I8P716 A0A2Z1UN50 B4G3T2 Q29A26 A0A3B0KPU8 A0A1L8EGH6 A0A2J7PWZ8 A0A1L8EGV8 A0A1L8EGP6 A0A1W4UQW7 B4R197 B4HFT6 D6WCN3 E2ADZ7 U3U9X8 A0A0A6Z9L3 A0A3L8DVG8
A0A1E1W802 A0A0L7LBS9 A0A0S1U0C3 A0A2A4J888 A0A2H1W587 A0A0C5BQ59 A0A194Q1M5 A0A0N0PCK1 A0A212ERE8 A0A1Y1LSL1 U5ELW0 B3P145 A0A1B6CDQ8 A0A1B0CG21 A0A1L8DNL3 A0A1L8D9Y1 A0A1L8DNM9 A0A1L8D9Z1 A0A1B0D9G5 A0A0L0CH44 B4PQ10 A0A1I8P716 A0A2Z1UN50 B4G3T2 Q29A26 A0A3B0KPU8 A0A1L8EGH6 A0A2J7PWZ8 A0A1L8EGV8 A0A1L8EGP6 A0A1W4UQW7 B4R197 B4HFT6 D6WCN3 E2ADZ7 U3U9X8 A0A0A6Z9L3 A0A3L8DVG8
Pubmed
EMBL
AB365354
BAG55002.1
KZ150268
PZC71698.1
KC340918
AGH25550.1
+ More
MH028075 AXY04244.1 MH028074 AXY04243.1 GDQN01007979 JAT83075.1 JTDY01001764 KOB72957.1 KT005960 RSAL01000052 ALM30311.1 RVE50192.1 NWSH01002737 PCG67603.1 ODYU01006393 SOQ48197.1 KJ783483 AJM76772.1 KQ459579 KPI99451.1 KQ460650 KPJ13115.1 AGBW02013025 OWR44070.1 GEZM01048037 JAV76652.1 GANO01004731 JAB55140.1 CH954181 EDV49234.1 GEDC01025725 JAS11573.1 AJWK01010652 AJWK01010653 AJWK01010654 AJWK01010655 GFDF01006026 JAV08058.1 GFDF01010806 JAV03278.1 GFDF01006025 JAV08059.1 GFDF01010805 JAV03279.1 AJVK01028222 AJVK01028223 JRES01000409 KNC31561.1 CM000160 EDW97237.1 KRK03602.1 KT222997 KT222998 ANT96533.1 ANT96534.1 CH479179 EDW25037.1 CM000070 EAL27525.2 KRT00087.1 KRT00088.1 OUUW01000013 SPP87866.1 GFDG01000967 JAV17832.1 NEVH01020866 PNF20850.1 GFDG01000966 JAV17833.1 GFDG01000965 JAV17834.1 CM000364 EDX13074.1 CH480815 EDW42321.1 KQ971317 EEZ97804.1 GL438827 EFN68230.1 AB817246 BAO00943.1 JQ866928 AFW19796.1 QOIP01000004 RLU23748.1
MH028075 AXY04244.1 MH028074 AXY04243.1 GDQN01007979 JAT83075.1 JTDY01001764 KOB72957.1 KT005960 RSAL01000052 ALM30311.1 RVE50192.1 NWSH01002737 PCG67603.1 ODYU01006393 SOQ48197.1 KJ783483 AJM76772.1 KQ459579 KPI99451.1 KQ460650 KPJ13115.1 AGBW02013025 OWR44070.1 GEZM01048037 JAV76652.1 GANO01004731 JAB55140.1 CH954181 EDV49234.1 GEDC01025725 JAS11573.1 AJWK01010652 AJWK01010653 AJWK01010654 AJWK01010655 GFDF01006026 JAV08058.1 GFDF01010806 JAV03278.1 GFDF01006025 JAV08059.1 GFDF01010805 JAV03279.1 AJVK01028222 AJVK01028223 JRES01000409 KNC31561.1 CM000160 EDW97237.1 KRK03602.1 KT222997 KT222998 ANT96533.1 ANT96534.1 CH479179 EDW25037.1 CM000070 EAL27525.2 KRT00087.1 KRT00088.1 OUUW01000013 SPP87866.1 GFDG01000967 JAV17832.1 NEVH01020866 PNF20850.1 GFDG01000966 JAV17833.1 GFDG01000965 JAV17834.1 CM000364 EDX13074.1 CH480815 EDW42321.1 KQ971317 EEZ97804.1 GL438827 EFN68230.1 AB817246 BAO00943.1 JQ866928 AFW19796.1 QOIP01000004 RLU23748.1
Proteomes
PRIDE
Interpro
IPR037729
CCHa1/2
ProteinModelPortal
B3XYE5
A0A2W1B8P0
C0HKT1
M4QFY3
A0A385H8U2
A0A385H8U4
+ More
A0A1E1W802 A0A0L7LBS9 A0A0S1U0C3 A0A2A4J888 A0A2H1W587 A0A0C5BQ59 A0A194Q1M5 A0A0N0PCK1 A0A212ERE8 A0A1Y1LSL1 U5ELW0 B3P145 A0A1B6CDQ8 A0A1B0CG21 A0A1L8DNL3 A0A1L8D9Y1 A0A1L8DNM9 A0A1L8D9Z1 A0A1B0D9G5 A0A0L0CH44 B4PQ10 A0A1I8P716 A0A2Z1UN50 B4G3T2 Q29A26 A0A3B0KPU8 A0A1L8EGH6 A0A2J7PWZ8 A0A1L8EGV8 A0A1L8EGP6 A0A1W4UQW7 B4R197 B4HFT6 D6WCN3 E2ADZ7 U3U9X8 A0A0A6Z9L3 A0A3L8DVG8
A0A1E1W802 A0A0L7LBS9 A0A0S1U0C3 A0A2A4J888 A0A2H1W587 A0A0C5BQ59 A0A194Q1M5 A0A0N0PCK1 A0A212ERE8 A0A1Y1LSL1 U5ELW0 B3P145 A0A1B6CDQ8 A0A1B0CG21 A0A1L8DNL3 A0A1L8D9Y1 A0A1L8DNM9 A0A1L8D9Z1 A0A1B0D9G5 A0A0L0CH44 B4PQ10 A0A1I8P716 A0A2Z1UN50 B4G3T2 Q29A26 A0A3B0KPU8 A0A1L8EGH6 A0A2J7PWZ8 A0A1L8EGV8 A0A1L8EGP6 A0A1W4UQW7 B4R197 B4HFT6 D6WCN3 E2ADZ7 U3U9X8 A0A0A6Z9L3 A0A3L8DVG8
Ontologies
PANTHER
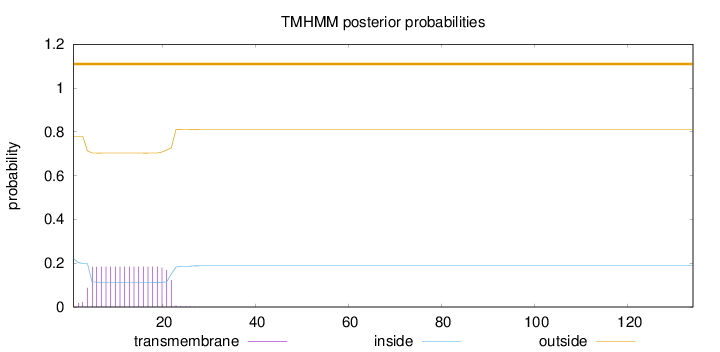
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 22,
Likelihood: 0.989265
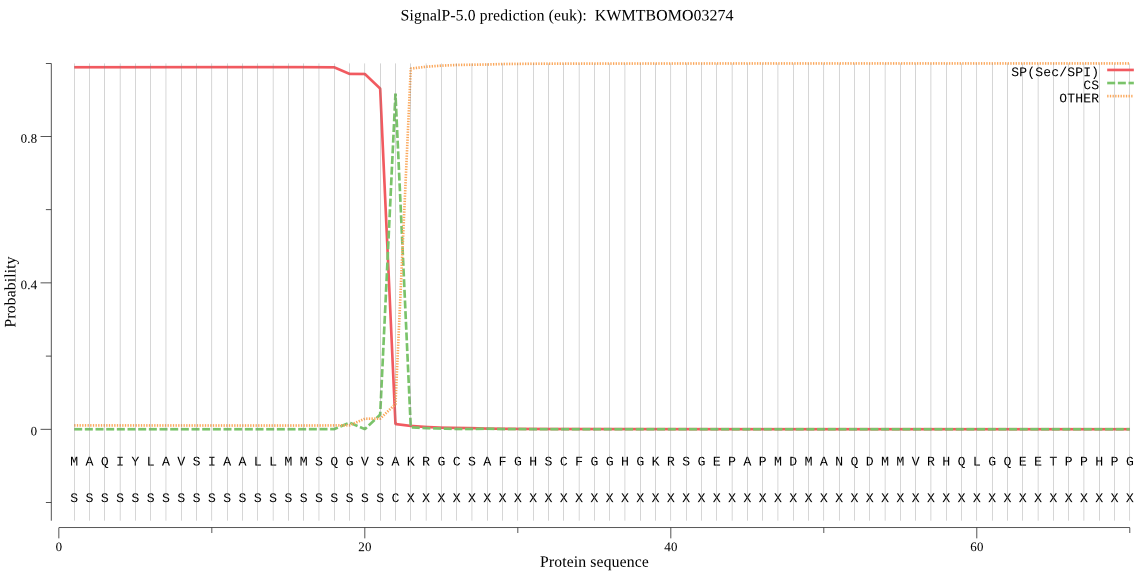
Length:
134
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.38402
Exp number, first 60 AAs:
3.38402
Total prob of N-in:
0.22118
outside
1 - 134
Population Genetic Test Statistics
Pi
20.211046
Theta
18.444009
Tajima's D
0.01141
CLR
0.357818
CSRT
0.376581170941453
Interpretation
Uncertain
