Gene
KWMTBOMO03272
Pre Gene Modal
BGIBMGA014557
Annotation
PREDICTED:_uncharacterized_protein_LOC101741188_[Bombyx_mori]
Full name
Phospholipase A(2)
Location in the cell
Extracellular Reliability : 2.085
Sequence
CDS
ATGTCCAATCCCCGCCTCAAACGAAACTTAAGTCAGGTTAGGTATATAACGAAGCCATACAAAAATCAAGTCAAAACTAGCAGTAGCAACCGTAGTACCTACGATGACACAGCCAAAGGGAACGAAATAGATTATAGGAAATTTTATAAAGATAAATCGAAACTGCTTGTCGGCTTCGCGAGAGATCCTTTCAGGTATTCAGTACCTCACGAGGATTACGCCAAGGGTGTGAGATACGTGAATCCTAATTCGTATAAAATGGATAATCGGGGTAAACGTGGTGTTTTCCACTTGTATAATATGATACAGTGCGCCACTGGATGTGAGCCTCTGTCGTATAAGGGGTATGGTTGTTATTGTGGATTTCTTGGAACTGGAAGACCCACAGATGGTATAGACAGGTGTTGCAAGATGCACGACCAGTGCTACGAGAATATCTACTGCCCCTTCTTCACGGTATACTTCCAGCCATACTACTGGAAGTGCTACAGAGGAGAGCCACTCTGCACTTTGGAGAACTATCAGAGTCAGGACCAATATGTTAATGGATGCGCCGCGAGACTTTGTGAATGCGACAGGTAA
Protein
MSNPRLKRNLSQVRYITKPYKNQVKTSSSNRSTYDDTAKGNEIDYRKFYKDKSKLLVGFARDPFRYSVPHEDYAKGVRYVNPNSYKMDNRGKRGVFHLYNMIQCATGCEPLSYKGYGCYCGFLGTGRPTDGIDRCCKMHDQCYENIYCPFFTVYFQPYYWKCYRGEPLCTLENYQSQDQYVNGCAARLCECDR
Summary
Catalytic Activity
a 1,2-diacyl-sn-glycero-3-phosphocholine + H2O = a 1-acyl-sn-glycero-3-phosphocholine + a fatty acid + H(+)
Cofactor
Ca(2+)
Similarity
Belongs to the phospholipase A2 family.
Feature
chain Phospholipase A(2)
Uniprot
A0A2W1BD03
A0A2H1WS96
A0A2H1WEK8
A0A212ERE3
A0A0L7KU97
A0A194Q291
+ More
I4DQF0 A0A0N1IP24 A0A0N0P9Q5 A0A2J7PWZ9 K7JAT1 A0A232EPW8 A0A067QH19 E2BCD7 A0A026W3T7 A0A182MQD2 A0A182JWP8 A0A182LAK5 A0A182SPQ5 A0A182VJQ1 A0A3L8DTW9 A0A1I8Q7A7 A0A182U9Y2 A0A1Y9J0R0 Q7QFU1 A0A310SPJ3 Q17F52 A0A0L0CQ08 A0A151I1R4 A0A1B6E0X3 A0A2S2QBD9 A0A1I8MLJ2 A0A0K8U7I2 A0A0K8UZE9 A0A0K8U8I6 A0A0J7L7K9 A0A336MN24 A0A336KH40 A0A2P8ZPM6 B3M1H9 B4HZ83 B4NFN2 B5DXT5 B4JYJ0 A0A1W4U633 B3DND2 Q8IML0 B4PQ48 A0A0M4EQA8 B4MBU6 B4K5X6 B3P5I9
I4DQF0 A0A0N1IP24 A0A0N0P9Q5 A0A2J7PWZ9 K7JAT1 A0A232EPW8 A0A067QH19 E2BCD7 A0A026W3T7 A0A182MQD2 A0A182JWP8 A0A182LAK5 A0A182SPQ5 A0A182VJQ1 A0A3L8DTW9 A0A1I8Q7A7 A0A182U9Y2 A0A1Y9J0R0 Q7QFU1 A0A310SPJ3 Q17F52 A0A0L0CQ08 A0A151I1R4 A0A1B6E0X3 A0A2S2QBD9 A0A1I8MLJ2 A0A0K8U7I2 A0A0K8UZE9 A0A0K8U8I6 A0A0J7L7K9 A0A336MN24 A0A336KH40 A0A2P8ZPM6 B3M1H9 B4HZ83 B4NFN2 B5DXT5 B4JYJ0 A0A1W4U633 B3DND2 Q8IML0 B4PQ48 A0A0M4EQA8 B4MBU6 B4K5X6 B3P5I9
EC Number
3.1.1.4
Pubmed
EMBL
KZ150268
PZC71694.1
ODYU01010668
SOQ55931.1
ODYU01008151
SOQ51520.1
+ More
AGBW02013025 OWR44073.1 JTDY01005757 KOB66665.1 KQ459579 KPI99453.1 AK404080 BAM20140.1 KQ460650 KPJ13119.1 KQ459510 KPJ00070.1 NEVH01020866 PNF20849.1 AAZX01001967 NNAY01002871 OXU20404.1 KK853452 KDR07539.1 GL447257 EFN86673.1 KK107453 EZA50663.1 AXCM01001896 QOIP01000004 RLU23746.1 AAAB01008844 EAA06034.4 KQ760539 OAD59987.1 CH477276 EAT45131.1 JRES01001062 JRES01000065 KNC25908.1 KNC34433.1 KQ976563 KYM80580.1 GEDC01005723 JAS31575.1 GGMS01005852 MBY75055.1 GDHF01030034 JAI22280.1 GDHF01020609 JAI31705.1 GDHF01029345 JAI22969.1 LBMM01000386 KMQ98641.1 UFQT01001807 SSX31844.1 UFQS01000417 UFQT01000417 SSX03759.1 SSX24124.1 PYGN01000002 PSN58423.1 CH902617 EDV43270.1 CH480819 EDW53340.1 CH964251 EDW83099.1 CM000070 EDY67526.2 CH916377 EDV90752.1 BT032920 ACD99484.1 AE014297 AAN14166.2 CM000160 EDW98317.1 CP012526 ALC47151.1 CH940656 EDW58567.1 CH933806 EDW16213.2 CH954182 EDV53239.1
AGBW02013025 OWR44073.1 JTDY01005757 KOB66665.1 KQ459579 KPI99453.1 AK404080 BAM20140.1 KQ460650 KPJ13119.1 KQ459510 KPJ00070.1 NEVH01020866 PNF20849.1 AAZX01001967 NNAY01002871 OXU20404.1 KK853452 KDR07539.1 GL447257 EFN86673.1 KK107453 EZA50663.1 AXCM01001896 QOIP01000004 RLU23746.1 AAAB01008844 EAA06034.4 KQ760539 OAD59987.1 CH477276 EAT45131.1 JRES01001062 JRES01000065 KNC25908.1 KNC34433.1 KQ976563 KYM80580.1 GEDC01005723 JAS31575.1 GGMS01005852 MBY75055.1 GDHF01030034 JAI22280.1 GDHF01020609 JAI31705.1 GDHF01029345 JAI22969.1 LBMM01000386 KMQ98641.1 UFQT01001807 SSX31844.1 UFQS01000417 UFQT01000417 SSX03759.1 SSX24124.1 PYGN01000002 PSN58423.1 CH902617 EDV43270.1 CH480819 EDW53340.1 CH964251 EDW83099.1 CM000070 EDY67526.2 CH916377 EDV90752.1 BT032920 ACD99484.1 AE014297 AAN14166.2 CM000160 EDW98317.1 CP012526 ALC47151.1 CH940656 EDW58567.1 CH933806 EDW16213.2 CH954182 EDV53239.1
Proteomes
UP000007151
UP000037510
UP000053268
UP000053240
UP000235965
UP000002358
+ More
UP000215335 UP000027135 UP000008237 UP000053097 UP000075883 UP000075881 UP000075882 UP000075901 UP000075903 UP000279307 UP000095300 UP000075902 UP000076407 UP000007062 UP000008820 UP000037069 UP000078540 UP000095301 UP000036403 UP000245037 UP000007801 UP000001292 UP000007798 UP000001819 UP000001070 UP000192221 UP000000803 UP000002282 UP000092553 UP000008792 UP000009192 UP000008711
UP000215335 UP000027135 UP000008237 UP000053097 UP000075883 UP000075881 UP000075882 UP000075901 UP000075903 UP000279307 UP000095300 UP000075902 UP000076407 UP000007062 UP000008820 UP000037069 UP000078540 UP000095301 UP000036403 UP000245037 UP000007801 UP000001292 UP000007798 UP000001819 UP000001070 UP000192221 UP000000803 UP000002282 UP000092553 UP000008792 UP000009192 UP000008711
PRIDE
Pfam
PF00068 Phospholip_A2_1
Interpro
SUPFAM
SSF48619
SSF48619
Gene 3D
CDD
ProteinModelPortal
A0A2W1BD03
A0A2H1WS96
A0A2H1WEK8
A0A212ERE3
A0A0L7KU97
A0A194Q291
+ More
I4DQF0 A0A0N1IP24 A0A0N0P9Q5 A0A2J7PWZ9 K7JAT1 A0A232EPW8 A0A067QH19 E2BCD7 A0A026W3T7 A0A182MQD2 A0A182JWP8 A0A182LAK5 A0A182SPQ5 A0A182VJQ1 A0A3L8DTW9 A0A1I8Q7A7 A0A182U9Y2 A0A1Y9J0R0 Q7QFU1 A0A310SPJ3 Q17F52 A0A0L0CQ08 A0A151I1R4 A0A1B6E0X3 A0A2S2QBD9 A0A1I8MLJ2 A0A0K8U7I2 A0A0K8UZE9 A0A0K8U8I6 A0A0J7L7K9 A0A336MN24 A0A336KH40 A0A2P8ZPM6 B3M1H9 B4HZ83 B4NFN2 B5DXT5 B4JYJ0 A0A1W4U633 B3DND2 Q8IML0 B4PQ48 A0A0M4EQA8 B4MBU6 B4K5X6 B3P5I9
I4DQF0 A0A0N1IP24 A0A0N0P9Q5 A0A2J7PWZ9 K7JAT1 A0A232EPW8 A0A067QH19 E2BCD7 A0A026W3T7 A0A182MQD2 A0A182JWP8 A0A182LAK5 A0A182SPQ5 A0A182VJQ1 A0A3L8DTW9 A0A1I8Q7A7 A0A182U9Y2 A0A1Y9J0R0 Q7QFU1 A0A310SPJ3 Q17F52 A0A0L0CQ08 A0A151I1R4 A0A1B6E0X3 A0A2S2QBD9 A0A1I8MLJ2 A0A0K8U7I2 A0A0K8UZE9 A0A0K8U8I6 A0A0J7L7K9 A0A336MN24 A0A336KH40 A0A2P8ZPM6 B3M1H9 B4HZ83 B4NFN2 B5DXT5 B4JYJ0 A0A1W4U633 B3DND2 Q8IML0 B4PQ48 A0A0M4EQA8 B4MBU6 B4K5X6 B3P5I9
PDB
1YXL
E-value=5.55381e-11,
Score=158
Ontologies
KEGG
PATHWAY
00564
Glycerophospholipid metabolism - Bombyx mori (domestic silkworm)
00565 Ether lipid metabolism - Bombyx mori (domestic silkworm)
00590 Arachidonic acid metabolism - Bombyx mori (domestic silkworm)
00591 Linoleic acid metabolism - Bombyx mori (domestic silkworm)
00592 alpha-Linolenic acid metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00565 Ether lipid metabolism - Bombyx mori (domestic silkworm)
00590 Arachidonic acid metabolism - Bombyx mori (domestic silkworm)
00591 Linoleic acid metabolism - Bombyx mori (domestic silkworm)
00592 alpha-Linolenic acid metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
PANTHER
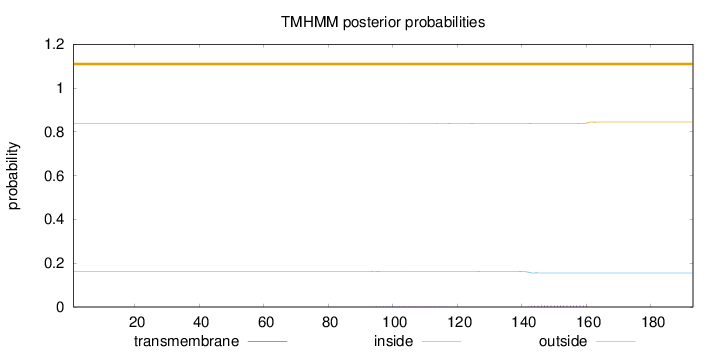
Topology
Subcellular location
Secreted
Length:
193
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15385
Exp number, first 60 AAs:
0
Total prob of N-in:
0.16332
outside
1 - 193
Population Genetic Test Statistics
Pi
179.366314
Theta
162.525638
Tajima's D
0.206233
CLR
0
CSRT
0.428578571071446
Interpretation
Uncertain
