Gene
KWMTBOMO03271 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014558
Annotation
PREDICTED:_CDK5_regulatory_subunit-associated_protein_3_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.818
Sequence
CDS
ATGGATGAAAGCAACATCCCCATCGATATAAACATAGGAAAGCTTCAAGACTGGTTGATCAGTCGTAGGCACGTCTCTAAAGATTGGCAGAAGAATGTCATAGCTGTGAGAGAGAAGATCAACAATGCCATCCAAGACATGCCGGCTCACCAAGACATAGCTGCTTTACTCTCAGGCAGCTACATAAATTATTTCCACTGCTTAAAAATCATTGAAATACTAAAAGAAACTGAGGCTGATACTAAAAATTTGTTCGGGAGATACGGGTCACAGAGGATGAAGGACTGGCAGGATGTTGTGAGGAACTATGAGAAGGAGAACTTGTATTTAGCAGAGGCAGCACAGATGCTGGTCAGGAATATTAGTTATGAGATTCCAGGGTTGAAGAGGCAGATTGCCAAAGAGGAACAGGCACAAGCGGACTTTGAAAAGAAGCATGCAGACTATCTAAAGAATGAAGCATCAAGTAAATCAGAGTTTCTGGCTTTGTGTAAACAACTTGGTATCCAAGGTGAGAAGATCAAGAGAGAATTAGTGGCGAAACTGCAGGAGCTGCCTGAGATATATGACCAAATAGGGAAATCACTAAAACCACTCCAGCCAGGTATAGAGTTATACAATGCATTCACAAAGTATATCCTCGGTGAGGATTCTCCAGAAGCTTTACCAATTTTGCAATATGTGATTATTCACGGCAATACTACAGTGTATGAATGGAGTTACGGAGAACCTCCTTTGAGCTTGGAGCCAGATCCTATACATATCGAATTAGATGATGACGAAGAACAGGCTTCTGGAGATCAGATAGATTTTGGGGACAACAATGATATAGATTTTTCTTCTATCGATGCTTCAGGCGCAGGGATTGACTTCGGAGATGGGGATGGGGCTGCTGAAATAGACTGGGGCAATATTGATATTGCTTCGGCCGATGAAAATAGTCTTTTGGCTGATATAGAAGCTGTGTCTTTGGAAGGGGCGGGGATAGTAATAGAGGAGCAAGGGATGACCGGTGGAGTTGCGAGGGGCAAAGATGCTCTGACGTTATTGGACAACCCCTCTACAAGGAACCAGTTCATCGATCAACTCGTTGAGCTCGAGTCGTTCCTGAAGATGCGCGTGTACGAGACGACGAGTGCGGCGGAGTCCCACACGCTGTCGCTGCTGCAGCAGCTGCCGGCGGAGAGCGAGCCGGCGCTGCGGGGCATGCTGGGCGCCGTGCAGCGCGCCGCCGCCGCCCTCGCCGCGCCCGACCTCGCGCACCTGCACAACGTCAAGCACTCGCCCAGGTACGTGGACGTGCTGACGGCTCAGCTAAAGCAGAAGCTGGTACTGTGCGAGAAGCTGTCGAAGCTGGCGGCGCGAGCGGCCGAACAGCGAACGGCGGCGAACGTTCGCGCCGCCGAACTAAGGCCGGTGCTCTCTAAAATCATAGAGAGAACTAAACAGTTGCAAGCGCAGATCGAAAGCGACATATCGAAGAAGTACAAGGGCAGGCCGGTCAACATTATCGGCGGTGTTAAATTCTTGTAG
Protein
MDESNIPIDINIGKLQDWLISRRHVSKDWQKNVIAVREKINNAIQDMPAHQDIAALLSGSYINYFHCLKIIEILKETEADTKNLFGRYGSQRMKDWQDVVRNYEKENLYLAEAAQMLVRNISYEIPGLKRQIAKEEQAQADFEKKHADYLKNEASSKSEFLALCKQLGIQGEKIKRELVAKLQELPEIYDQIGKSLKPLQPGIELYNAFTKYILGEDSPEALPILQYVIIHGNTTVYEWSYGEPPLSLEPDPIHIELDDDEEQASGDQIDFGDNNDIDFSSIDASGAGIDFGDGDGAAEIDWGNIDIASADENSLLADIEAVSLEGAGIVIEEQGMTGGVARGKDALTLLDNPSTRNQFIDQLVELESFLKMRVYETTSAAESHTLSLLQQLPAESEPALRGMLGAVQRAAAALAAPDLAHLHNVKHSPRYVDVLTAQLKQKLVLCEKLSKLAARAAEQRTAANVRAAELRPVLSKIIERTKQLQAQIESDISKKYKGRPVNIIGGVKFL
Summary
Uniprot
A0A2W1B6M7
A0A2H1WWN3
A0A0N1PGX6
A0A212ERJ5
H9JYD8
A0A194Q374
+ More
A0A0T6B2R8 D6WFV3 A0A1W4XIP0 A0A2J7PWZ1 A0A087ZP88 A0A154P7E1 A0A310SS64 A0A1Y1MDJ6 A0A2A3ESJ9 K7JAT0 A0A0J7L7E2 A0A0N0BH63 A0A232EPW1 N6U9G8 U4U1C0 A0A1B6ES14 A0A1L8DM00 A0A1B0GNA4 F4W5Z9 B0W1D5 Q17IE6 A0A1B6HSQ6 A0A1B6L5Z9 A0A182H4N0 A0A026W6L4 A0A2M3Z7J5 U5EMN2 A0A182SQY6 A0A1Q3FBF7 A0A1Q3FBV0 A0A2M4BLP3 A0A2M4BKW0 A0A151JPE6 A0A2M4BKV8 A0A182YDH1 A0A3L8DTJ9 A0A0P5DXL4 A0A1B0CS87 A0A0L7R2I5 W5J450 E9G1K1 A0A1B6CFW3 A0A182RQ61 A0A182QMX8 E2BCD6 A0A182F1Q3 A0A182KTV7 A0A084WG45 A0A182XGC4 A0A182V555 A0A2C9GRA9 Q7Q1F7 A0A0P6BVL4 A0A1J1HHS6 A0A1B6IFW6 A0A182K6P7 A0A182ITW4 A0A2M4AAM0 A0A182UF71 A0A182MJF7 A0A067QGR3 A0A0P6JJI0 A0A0K8TRB8 A0A182NBZ5 A0A336LTA9 A0A131XEB8 A0A0P6CWY5 A0A131YUT6 A0A151X4P4 A0A0P5LRE5 A0A182P322 A0A151JY84 G3MLF3 A0A023ESU5 A0A182WGP4 A0A224YQA4 E2ADZ9 A0A1I8N5H6 A0A195CU05 A0A1I8N5I1 A0A1E1XCR4 A0A1L8DLC9 L7LUV6 A0A1A9X2Z2 A0A131Y046 V5HXS5 A0A1S3JUE8 A0A0P4VRI4 A0A1B0G2F0 A0A293N0Z6 D3TL28 A0A1A9V8M1 A0A1A9ZVI5
A0A0T6B2R8 D6WFV3 A0A1W4XIP0 A0A2J7PWZ1 A0A087ZP88 A0A154P7E1 A0A310SS64 A0A1Y1MDJ6 A0A2A3ESJ9 K7JAT0 A0A0J7L7E2 A0A0N0BH63 A0A232EPW1 N6U9G8 U4U1C0 A0A1B6ES14 A0A1L8DM00 A0A1B0GNA4 F4W5Z9 B0W1D5 Q17IE6 A0A1B6HSQ6 A0A1B6L5Z9 A0A182H4N0 A0A026W6L4 A0A2M3Z7J5 U5EMN2 A0A182SQY6 A0A1Q3FBF7 A0A1Q3FBV0 A0A2M4BLP3 A0A2M4BKW0 A0A151JPE6 A0A2M4BKV8 A0A182YDH1 A0A3L8DTJ9 A0A0P5DXL4 A0A1B0CS87 A0A0L7R2I5 W5J450 E9G1K1 A0A1B6CFW3 A0A182RQ61 A0A182QMX8 E2BCD6 A0A182F1Q3 A0A182KTV7 A0A084WG45 A0A182XGC4 A0A182V555 A0A2C9GRA9 Q7Q1F7 A0A0P6BVL4 A0A1J1HHS6 A0A1B6IFW6 A0A182K6P7 A0A182ITW4 A0A2M4AAM0 A0A182UF71 A0A182MJF7 A0A067QGR3 A0A0P6JJI0 A0A0K8TRB8 A0A182NBZ5 A0A336LTA9 A0A131XEB8 A0A0P6CWY5 A0A131YUT6 A0A151X4P4 A0A0P5LRE5 A0A182P322 A0A151JY84 G3MLF3 A0A023ESU5 A0A182WGP4 A0A224YQA4 E2ADZ9 A0A1I8N5H6 A0A195CU05 A0A1I8N5I1 A0A1E1XCR4 A0A1L8DLC9 L7LUV6 A0A1A9X2Z2 A0A131Y046 V5HXS5 A0A1S3JUE8 A0A0P4VRI4 A0A1B0G2F0 A0A293N0Z6 D3TL28 A0A1A9V8M1 A0A1A9ZVI5
Pubmed
28756777
26354079
22118469
19121390
18362917
19820115
+ More
28004739 20075255 28648823 23537049 21719571 17510324 26483478 24508170 25244985 30249741 20920257 23761445 21292972 20798317 20966253 24438588 12364791 14747013 17210077 24845553 26369729 28049606 26830274 22216098 24945155 28797301 25315136 28503490 25576852 25765539 20353571
28004739 20075255 28648823 23537049 21719571 17510324 26483478 24508170 25244985 30249741 20920257 23761445 21292972 20798317 20966253 24438588 12364791 14747013 17210077 24845553 26369729 28049606 26830274 22216098 24945155 28797301 25315136 28503490 25576852 25765539 20353571
EMBL
KZ150268
PZC71692.1
ODYU01011560
SOQ57376.1
KQ460650
KPJ13120.1
+ More
AGBW02013025 OWR44074.1 BABH01043417 BABH01043418 KQ459579 KPI99454.1 LJIG01016080 KRT81674.1 KQ971328 EFA00941.1 NEVH01020866 PNF20855.1 KQ434829 KZC07753.1 KQ760539 OAD59986.1 GEZM01036470 JAV82670.1 KZ288186 PBC34763.1 AAZX01001967 LBMM01000386 KMQ98640.1 KQ435758 KOX75784.1 NNAY01002871 OXU20405.1 APGK01034486 APGK01034487 KB740914 ENN78335.1 KB631869 ERL86842.1 GECZ01029069 JAS40700.1 GFDF01006598 JAV07486.1 AJVK01013092 GL887696 EGI70377.1 DS231821 EDS45005.1 CH477240 EAT46431.1 GECU01030010 JAS77696.1 GEBQ01020902 JAT19075.1 JXUM01109896 KQ565364 KXJ71049.1 KK107453 EZA50664.1 GGFM01003709 MBW24460.1 GANO01000949 JAB58922.1 GFDL01010155 JAV24890.1 GFDL01010072 JAV24973.1 GGFJ01004567 MBW53708.1 GGFJ01004566 MBW53707.1 KQ978737 KYN28904.1 GGFJ01004565 MBW53706.1 QOIP01000004 RLU23747.1 GDIP01149944 GDIQ01034959 JAJ73458.1 JAN59778.1 AJWK01025802 AJWK01025803 KQ414666 KOC65049.1 ADMH02002127 ETN58631.1 GL732529 EFX86797.1 GEDC01024922 JAS12376.1 AXCN02000692 GL447257 EFN86672.1 ATLV01023440 KE525343 KFB49189.1 APCN01001736 AAAB01008980 EAA14226.4 GDIP01009038 JAM94677.1 CVRI01000004 CRK87443.1 GECU01021892 JAS85814.1 GGFK01004513 MBW37834.1 AXCM01001183 KK853452 KDR07538.1 GDIQ01005594 JAN89143.1 GDAI01000684 JAI16919.1 UFQT01000176 SSX21160.1 GEFH01004595 JAP63986.1 GDIQ01088840 JAN05897.1 GEDV01005568 JAP82989.1 KQ982543 KYQ55393.1 GDIQ01167069 JAK84656.1 KQ981505 KYN40920.1 JO842704 AEO34321.1 GAPW01001235 JAC12363.1 GFPF01005038 MAA16184.1 GL438827 EFN68232.1 KQ977306 KYN03614.1 GFAC01002190 JAT96998.1 GFDF01006832 JAV07252.1 GACK01010361 JAA54673.1 GEFM01003157 JAP72639.1 GANP01000939 JAB83529.1 GDRN01104755 GDRN01104753 JAI57863.1 CCAG010006799 GFWV01021331 MAA46059.1 EZ422124 ADD18323.1
AGBW02013025 OWR44074.1 BABH01043417 BABH01043418 KQ459579 KPI99454.1 LJIG01016080 KRT81674.1 KQ971328 EFA00941.1 NEVH01020866 PNF20855.1 KQ434829 KZC07753.1 KQ760539 OAD59986.1 GEZM01036470 JAV82670.1 KZ288186 PBC34763.1 AAZX01001967 LBMM01000386 KMQ98640.1 KQ435758 KOX75784.1 NNAY01002871 OXU20405.1 APGK01034486 APGK01034487 KB740914 ENN78335.1 KB631869 ERL86842.1 GECZ01029069 JAS40700.1 GFDF01006598 JAV07486.1 AJVK01013092 GL887696 EGI70377.1 DS231821 EDS45005.1 CH477240 EAT46431.1 GECU01030010 JAS77696.1 GEBQ01020902 JAT19075.1 JXUM01109896 KQ565364 KXJ71049.1 KK107453 EZA50664.1 GGFM01003709 MBW24460.1 GANO01000949 JAB58922.1 GFDL01010155 JAV24890.1 GFDL01010072 JAV24973.1 GGFJ01004567 MBW53708.1 GGFJ01004566 MBW53707.1 KQ978737 KYN28904.1 GGFJ01004565 MBW53706.1 QOIP01000004 RLU23747.1 GDIP01149944 GDIQ01034959 JAJ73458.1 JAN59778.1 AJWK01025802 AJWK01025803 KQ414666 KOC65049.1 ADMH02002127 ETN58631.1 GL732529 EFX86797.1 GEDC01024922 JAS12376.1 AXCN02000692 GL447257 EFN86672.1 ATLV01023440 KE525343 KFB49189.1 APCN01001736 AAAB01008980 EAA14226.4 GDIP01009038 JAM94677.1 CVRI01000004 CRK87443.1 GECU01021892 JAS85814.1 GGFK01004513 MBW37834.1 AXCM01001183 KK853452 KDR07538.1 GDIQ01005594 JAN89143.1 GDAI01000684 JAI16919.1 UFQT01000176 SSX21160.1 GEFH01004595 JAP63986.1 GDIQ01088840 JAN05897.1 GEDV01005568 JAP82989.1 KQ982543 KYQ55393.1 GDIQ01167069 JAK84656.1 KQ981505 KYN40920.1 JO842704 AEO34321.1 GAPW01001235 JAC12363.1 GFPF01005038 MAA16184.1 GL438827 EFN68232.1 KQ977306 KYN03614.1 GFAC01002190 JAT96998.1 GFDF01006832 JAV07252.1 GACK01010361 JAA54673.1 GEFM01003157 JAP72639.1 GANP01000939 JAB83529.1 GDRN01104755 GDRN01104753 JAI57863.1 CCAG010006799 GFWV01021331 MAA46059.1 EZ422124 ADD18323.1
Proteomes
UP000053240
UP000007151
UP000005204
UP000053268
UP000007266
UP000192223
+ More
UP000235965 UP000005203 UP000076502 UP000242457 UP000002358 UP000036403 UP000053105 UP000215335 UP000019118 UP000030742 UP000092462 UP000007755 UP000002320 UP000008820 UP000069940 UP000249989 UP000053097 UP000075901 UP000078492 UP000076408 UP000279307 UP000092461 UP000053825 UP000000673 UP000000305 UP000075900 UP000075886 UP000008237 UP000069272 UP000075882 UP000030765 UP000076407 UP000075903 UP000075840 UP000007062 UP000183832 UP000075881 UP000075880 UP000075902 UP000075883 UP000027135 UP000075884 UP000075809 UP000075885 UP000078541 UP000075920 UP000000311 UP000095301 UP000078542 UP000091820 UP000085678 UP000092444 UP000078200 UP000092445
UP000235965 UP000005203 UP000076502 UP000242457 UP000002358 UP000036403 UP000053105 UP000215335 UP000019118 UP000030742 UP000092462 UP000007755 UP000002320 UP000008820 UP000069940 UP000249989 UP000053097 UP000075901 UP000078492 UP000076408 UP000279307 UP000092461 UP000053825 UP000000673 UP000000305 UP000075900 UP000075886 UP000008237 UP000069272 UP000075882 UP000030765 UP000076407 UP000075903 UP000075840 UP000007062 UP000183832 UP000075881 UP000075880 UP000075902 UP000075883 UP000027135 UP000075884 UP000075809 UP000075885 UP000078541 UP000075920 UP000000311 UP000095301 UP000078542 UP000091820 UP000085678 UP000092444 UP000078200 UP000092445
PRIDE
SUPFAM
SSF101576
SSF101576
ProteinModelPortal
A0A2W1B6M7
A0A2H1WWN3
A0A0N1PGX6
A0A212ERJ5
H9JYD8
A0A194Q374
+ More
A0A0T6B2R8 D6WFV3 A0A1W4XIP0 A0A2J7PWZ1 A0A087ZP88 A0A154P7E1 A0A310SS64 A0A1Y1MDJ6 A0A2A3ESJ9 K7JAT0 A0A0J7L7E2 A0A0N0BH63 A0A232EPW1 N6U9G8 U4U1C0 A0A1B6ES14 A0A1L8DM00 A0A1B0GNA4 F4W5Z9 B0W1D5 Q17IE6 A0A1B6HSQ6 A0A1B6L5Z9 A0A182H4N0 A0A026W6L4 A0A2M3Z7J5 U5EMN2 A0A182SQY6 A0A1Q3FBF7 A0A1Q3FBV0 A0A2M4BLP3 A0A2M4BKW0 A0A151JPE6 A0A2M4BKV8 A0A182YDH1 A0A3L8DTJ9 A0A0P5DXL4 A0A1B0CS87 A0A0L7R2I5 W5J450 E9G1K1 A0A1B6CFW3 A0A182RQ61 A0A182QMX8 E2BCD6 A0A182F1Q3 A0A182KTV7 A0A084WG45 A0A182XGC4 A0A182V555 A0A2C9GRA9 Q7Q1F7 A0A0P6BVL4 A0A1J1HHS6 A0A1B6IFW6 A0A182K6P7 A0A182ITW4 A0A2M4AAM0 A0A182UF71 A0A182MJF7 A0A067QGR3 A0A0P6JJI0 A0A0K8TRB8 A0A182NBZ5 A0A336LTA9 A0A131XEB8 A0A0P6CWY5 A0A131YUT6 A0A151X4P4 A0A0P5LRE5 A0A182P322 A0A151JY84 G3MLF3 A0A023ESU5 A0A182WGP4 A0A224YQA4 E2ADZ9 A0A1I8N5H6 A0A195CU05 A0A1I8N5I1 A0A1E1XCR4 A0A1L8DLC9 L7LUV6 A0A1A9X2Z2 A0A131Y046 V5HXS5 A0A1S3JUE8 A0A0P4VRI4 A0A1B0G2F0 A0A293N0Z6 D3TL28 A0A1A9V8M1 A0A1A9ZVI5
A0A0T6B2R8 D6WFV3 A0A1W4XIP0 A0A2J7PWZ1 A0A087ZP88 A0A154P7E1 A0A310SS64 A0A1Y1MDJ6 A0A2A3ESJ9 K7JAT0 A0A0J7L7E2 A0A0N0BH63 A0A232EPW1 N6U9G8 U4U1C0 A0A1B6ES14 A0A1L8DM00 A0A1B0GNA4 F4W5Z9 B0W1D5 Q17IE6 A0A1B6HSQ6 A0A1B6L5Z9 A0A182H4N0 A0A026W6L4 A0A2M3Z7J5 U5EMN2 A0A182SQY6 A0A1Q3FBF7 A0A1Q3FBV0 A0A2M4BLP3 A0A2M4BKW0 A0A151JPE6 A0A2M4BKV8 A0A182YDH1 A0A3L8DTJ9 A0A0P5DXL4 A0A1B0CS87 A0A0L7R2I5 W5J450 E9G1K1 A0A1B6CFW3 A0A182RQ61 A0A182QMX8 E2BCD6 A0A182F1Q3 A0A182KTV7 A0A084WG45 A0A182XGC4 A0A182V555 A0A2C9GRA9 Q7Q1F7 A0A0P6BVL4 A0A1J1HHS6 A0A1B6IFW6 A0A182K6P7 A0A182ITW4 A0A2M4AAM0 A0A182UF71 A0A182MJF7 A0A067QGR3 A0A0P6JJI0 A0A0K8TRB8 A0A182NBZ5 A0A336LTA9 A0A131XEB8 A0A0P6CWY5 A0A131YUT6 A0A151X4P4 A0A0P5LRE5 A0A182P322 A0A151JY84 G3MLF3 A0A023ESU5 A0A182WGP4 A0A224YQA4 E2ADZ9 A0A1I8N5H6 A0A195CU05 A0A1I8N5I1 A0A1E1XCR4 A0A1L8DLC9 L7LUV6 A0A1A9X2Z2 A0A131Y046 V5HXS5 A0A1S3JUE8 A0A0P4VRI4 A0A1B0G2F0 A0A293N0Z6 D3TL28 A0A1A9V8M1 A0A1A9ZVI5
Ontologies
PANTHER
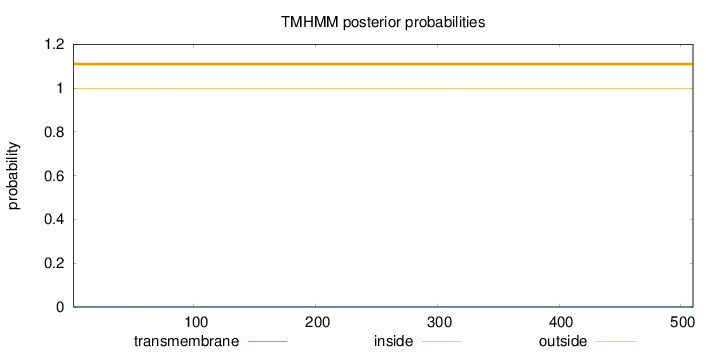
Topology
Length:
510
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00339
Exp number, first 60 AAs:
0.0013
Total prob of N-in:
0.00131
outside
1 - 510
Population Genetic Test Statistics
Pi
337.450159
Theta
173.171113
Tajima's D
2.699277
CLR
0.489625
CSRT
0.959302034898255
Interpretation
Uncertain
