Gene
KWMTBOMO03269
Annotation
PREDICTED:_peroxidase-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.337
Sequence
CDS
ATGCAATGGGGTCAATTTATCACTCATGACATGGTCTCTAAAGCTATGGAAGTCACTGACGAAGGCGGTATACAGTGTTGTCTTGGTGACGGCCAAGGAGTCCTACCACCGGAGTGGCTGAACGACAAATGCATACCGATCAGTGTGCCCGACGACGACACATTCTACCGGTCTTATGGCATTAAGTGCTTGAATATGGTGCGCAGTATCACCGCTCCAAGAGAAGACTGTACATTAGGACCCGCTGAACAGATGAACGCAGTGACCAGTTTCCTAGACGGCTCAACGATTTACGGCTCGGACAAGAAAACCTCAATGAAGCTTAGAAGCAAGAAAAACGGGAAACTTCGACAAGTCAACAAGAAGGACTCTTGTCGGGGATTTCTCCCGTCGGTCGACGACAAATCGGCCTCATGCGACTTGAGGAACTCCTCGGAGCCGTGCTACGTTTCAGGTGACCTCCGTGTAAACCAAACTCCCACCTTGGCCATGCTGCACACACTATTCCTGAGAGAGCACAACAGGATAGCGGATATTTTAAGCGAACTAAACCCGCTGTGGAGCGACGAAAAACTGTATCAAGAGACGAGAAAGATCGTTATAGCCGAAATACAACATATCACTTACCAGGAATGGCTGCCGGCTAATTTAGGTGAGAGCTACGTTCGATATTACCGCATATCTCCGTCGAGCCTTTACAGCAGAGACTACAACGAGGACGTGAATCCCGGTATTATCAATAGCTTCGGAGCGGCCGCATTTCGTTTCATGCACTCTGTCGTACCGGATACGATCATGGCCTGTCCCTCTGGCTATAATGCCGCCTATTTACACAGATTAAGTGACCACTATTTTAACCCAAGCATTCTAGAAACATCTTCGGAGGCGTTCGACGATGTCGTCAGAGGCTCAGTGAGTCATTACTCGGCCGAGGCGGACCCTTTTGTCAGTGGCGAATTAACTAATTTAATGTTTAAATCGTATAATCGATGGGGTCTGGATTTGATTGCTACGGATATACAGCGAGGACGGGACCATGGGCTGGCTTCTTATAACGATTACAGGGAAATCTGTGGCCTTCGAAAGGCGAGGTGTTTCCAAGATTTAGCTGGAGAAATCACACAAGACCGTATCAACGCCCTGGCCCGTCTCTACGAGTGCGTGGACGACATAGACTTGTTTGTCGGCGGATCTCTGGAACGCGACGTTCAGGGTTCGATATTCGGTCACACTTTTCACTGCATAGTGGCTGAACAGTTCTACAGAACGAGAATCGGCGATAGATTCTTCTATGACAACGGAGAAATGCCTCATTCTTTTAGCTCAGATCAACTGAAAGAAATAAAGAAAACTTCTCTGGCCCGTGTAATTTGCGACAACACAGACGGAGTCTACTACGTTCAGAAGAAGGCCTTCGAGCTGGAATCTACGTACAACCCCAAATATAGATGCGACGACTATGAGGCCATACCTTACGTGGATCTGACTGCGTGGAAGAAACCAATTTTAGATTTGTTCGATTGA
Protein
MQWGQFITHDMVSKAMEVTDEGGIQCCLGDGQGVLPPEWLNDKCIPISVPDDDTFYRSYGIKCLNMVRSITAPREDCTLGPAEQMNAVTSFLDGSTIYGSDKKTSMKLRSKKNGKLRQVNKKDSCRGFLPSVDDKSASCDLRNSSEPCYVSGDLRVNQTPTLAMLHTLFLREHNRIADILSELNPLWSDEKLYQETRKIVIAEIQHITYQEWLPANLGESYVRYYRISPSSLYSRDYNEDVNPGIINSFGAAAFRFMHSVVPDTIMACPSGYNAAYLHRLSDHYFNPSILETSSEAFDDVVRGSVSHYSAEADPFVSGELTNLMFKSYNRWGLDLIATDIQRGRDHGLASYNDYREICGLRKARCFQDLAGEITQDRINALARLYECVDDIDLFVGGSLERDVQGSIFGHTFHCIVAEQFYRTRIGDRFFYDNGEMPHSFSSDQLKEIKKTSLARVICDNTDGVYYVQKKAFELESTYNPKYRCDDYEAIPYVDLTAWKKPILDLFD
Summary
Uniprot
A0A194Q1N0
A0A212ERF1
A0A2W1BFY1
A0A2H1WEE0
A0A1B6MDD3
A0A232F238
+ More
K7IYZ2 A0A1B6CUT2 A0A2S2R1E6 A0A2H8TD85 A0A0P4VM15 X1WIY8 A0A146M0F8 T1I7W3 E9IRB0 A0A0J7KKC2 A0A0P5K7G1 E2ADZ2 A0A0P5LUG0 A0A0K8T8R0 A0A0P5M7P6 A0A0P6DES3 A0A1B6DAS3 A0A0N8EE02 A0A2P2IA20 A0A0P5SF47 A0A0N8C9P0 A0A2P8ZI40 A0A1B6CHC4 A0A0P5CJI8 A0A158NBE1 A0A0P5G0R9 A0A0P5I633 A0A0A9ZAS3 A0A162Q098 A0A0N8EED2 A0A195BAD8 A0A0P4ZMN0 T1HAC7 A0A0P6EWY1 A0A0P5Y961 A0A2A3EU99 A0A195DRC5 A0A0P6E070 A0A162Q0A9 V9IAE9 A0A0N8CUK9 V9ICS7 A0A2A3E974 A0A2J7RAN0 A0A0P5ULQ4 A0A0P5RU22 A0A226DGH2 A0A088AE09 A0A2J7RAL9 A0A0P6AZR4 A0A0N8BFE3 A0A0P5URI8 A0A0P5TIN6 A0A0N8DGW9 A0A0N8ENK5 A0A195FC13 A0A0P6JHY0 A0A0P5DSD1 A0A0A9Z8J0 F4X8U9 A0A0N8CQ89 A0A0P5AI81 A0A0P5PVA2 A0A0P6FQD9 A0A0P5BHY2 A0A0P5KKX2 A0A0P5UR53 A0A0N8A457 A0A0P5VCX6 A0A0P5AMC4 A0A0P5SG58 A0A0P5Q175 A0A0N7ZU83 A0A088AE08 A0A0P5XPB9 A0A0P5U773 A0A0P5AWG5 A0A0P6G880 A0A0P5ULU2 A0A146L8Z9 A0A0P5SEE6 A0A089FQD2 A0A0P5RV48 A0A0P5QGH8 A0A0N8E0L1 A0A0C9R1B3 A0A3L8DTP5 A0A0P6DHJ3 A0A0P5GUL0 A0A0P5BQS1 A0A0P6D9I9 A0A0P5BVJ2
K7IYZ2 A0A1B6CUT2 A0A2S2R1E6 A0A2H8TD85 A0A0P4VM15 X1WIY8 A0A146M0F8 T1I7W3 E9IRB0 A0A0J7KKC2 A0A0P5K7G1 E2ADZ2 A0A0P5LUG0 A0A0K8T8R0 A0A0P5M7P6 A0A0P6DES3 A0A1B6DAS3 A0A0N8EE02 A0A2P2IA20 A0A0P5SF47 A0A0N8C9P0 A0A2P8ZI40 A0A1B6CHC4 A0A0P5CJI8 A0A158NBE1 A0A0P5G0R9 A0A0P5I633 A0A0A9ZAS3 A0A162Q098 A0A0N8EED2 A0A195BAD8 A0A0P4ZMN0 T1HAC7 A0A0P6EWY1 A0A0P5Y961 A0A2A3EU99 A0A195DRC5 A0A0P6E070 A0A162Q0A9 V9IAE9 A0A0N8CUK9 V9ICS7 A0A2A3E974 A0A2J7RAN0 A0A0P5ULQ4 A0A0P5RU22 A0A226DGH2 A0A088AE09 A0A2J7RAL9 A0A0P6AZR4 A0A0N8BFE3 A0A0P5URI8 A0A0P5TIN6 A0A0N8DGW9 A0A0N8ENK5 A0A195FC13 A0A0P6JHY0 A0A0P5DSD1 A0A0A9Z8J0 F4X8U9 A0A0N8CQ89 A0A0P5AI81 A0A0P5PVA2 A0A0P6FQD9 A0A0P5BHY2 A0A0P5KKX2 A0A0P5UR53 A0A0N8A457 A0A0P5VCX6 A0A0P5AMC4 A0A0P5SG58 A0A0P5Q175 A0A0N7ZU83 A0A088AE08 A0A0P5XPB9 A0A0P5U773 A0A0P5AWG5 A0A0P6G880 A0A0P5ULU2 A0A146L8Z9 A0A0P5SEE6 A0A089FQD2 A0A0P5RV48 A0A0P5QGH8 A0A0N8E0L1 A0A0C9R1B3 A0A3L8DTP5 A0A0P6DHJ3 A0A0P5GUL0 A0A0P5BQS1 A0A0P6D9I9 A0A0P5BVJ2
Pubmed
EMBL
KQ459579
KPI99456.1
AGBW02013025
OWR44076.1
KZ150268
PZC71690.1
+ More
ODYU01008112 SOQ51433.1 GEBQ01006041 JAT33936.1 NNAY01001189 OXU24804.1 GEDC01020121 JAS17177.1 GGMS01014387 MBY83590.1 GFXV01000266 MBW12071.1 GDKW01003332 JAI53263.1 ABLF02037950 ABLF02037951 GDHC01005325 JAQ13304.1 ACPB03011632 GL765107 EFZ16876.1 LBMM01006170 KMQ90863.1 GDIQ01188338 JAK63387.1 GL438827 EFN68225.1 GDIQ01189639 JAK62086.1 GBRD01012786 GBRD01004319 GBRD01004311 JAG61510.1 GDIQ01171501 JAK80224.1 GDIQ01081745 JAN12992.1 GEDC01030510 GEDC01015160 GEDC01014578 GEDC01014226 GEDC01012292 GEDC01011530 JAS06788.1 JAS22138.1 JAS22720.1 JAS23072.1 JAS25006.1 JAS25768.1 GDIQ01036052 JAN58685.1 IACF01005278 LAB70864.1 GDIP01140921 JAL62793.1 GDIQ01101139 JAL50587.1 PYGN01000050 PSN56140.1 GEDC01029182 GEDC01024486 GEDC01016035 JAS08116.1 JAS12812.1 JAS21263.1 GDIP01169663 JAJ53739.1 ADTU01010985 ADTU01010986 ADTU01010987 GDIQ01248330 JAK03395.1 GDIQ01217481 JAK34244.1 GBHO01004699 JAG38905.1 LRGB01000389 KZS19289.1 GDIQ01035012 JAN59725.1 KQ976540 KYM81185.1 GDIP01210375 JAJ13027.1 ACPB03014183 GDIQ01069489 JAN25248.1 GDIP01062691 JAM41024.1 KZ288187 PBC34719.1 KQ980581 KYN15465.1 GDIQ01084694 JAN10043.1 KZS19290.1 JR038044 AEY58093.1 GDIP01105282 GDIP01097375 JAM06340.1 JR038045 AEY58094.1 KZ288315 PBC28333.1 NEVH01006567 PNF37882.1 GDIP01111181 JAL92533.1 GDIQ01096230 JAL55496.1 LNIX01000021 OXA43787.1 PNF37881.1 GDIP01022054 JAM81661.1 GDIQ01183115 JAK68610.1 GDIP01109537 JAL94177.1 GDIP01131493 JAL72221.1 GDIP01034895 JAM68820.1 GDIQ01009228 JAN85509.1 KQ981673 KYN38170.1 GDIQ01007615 JAN87122.1 GDIP01152383 JAJ71019.1 GBHO01042701 GBHO01003941 JAG00903.1 JAG39663.1 GL888932 EGI57368.1 GDIP01109535 JAL94179.1 GDIP01212074 JAJ11328.1 GDIQ01123725 JAL28001.1 GDIQ01055811 JAN38926.1 GDIP01184280 JAJ39122.1 GDIQ01184049 GDIP01134043 JAK67676.1 JAL69671.1 GDIP01109536 JAL94178.1 GDIP01184281 JAJ39121.1 GDIP01101490 JAM02225.1 GDIP01196765 JAJ26637.1 GDIQ01096228 JAL55498.1 GDIQ01121174 JAL30552.1 GDIP01212073 GDIQ01187448 GDIQ01123724 JAJ11329.1 JAK64277.1 GDIP01069438 JAM34277.1 GDIP01134089 JAL69625.1 GDIP01193179 JAJ30223.1 GDIQ01055810 JAN38927.1 GDIP01111182 JAL92532.1 GDHC01015042 JAQ03587.1 GDIQ01105580 JAL46146.1 KM025027 AIP87047.1 GDIQ01096229 JAL55497.1 GDIQ01121173 JAL30553.1 GDIQ01175304 GDIQ01073580 JAN21157.1 GBYB01000656 JAG70423.1 QOIP01000004 RLU23754.1 GDIQ01077337 JAN17400.1 GDIQ01239538 JAK12187.1 GDIP01181228 JAJ42174.1 GDIQ01082235 JAN12502.1 GDIP01179325 JAJ44077.1
ODYU01008112 SOQ51433.1 GEBQ01006041 JAT33936.1 NNAY01001189 OXU24804.1 GEDC01020121 JAS17177.1 GGMS01014387 MBY83590.1 GFXV01000266 MBW12071.1 GDKW01003332 JAI53263.1 ABLF02037950 ABLF02037951 GDHC01005325 JAQ13304.1 ACPB03011632 GL765107 EFZ16876.1 LBMM01006170 KMQ90863.1 GDIQ01188338 JAK63387.1 GL438827 EFN68225.1 GDIQ01189639 JAK62086.1 GBRD01012786 GBRD01004319 GBRD01004311 JAG61510.1 GDIQ01171501 JAK80224.1 GDIQ01081745 JAN12992.1 GEDC01030510 GEDC01015160 GEDC01014578 GEDC01014226 GEDC01012292 GEDC01011530 JAS06788.1 JAS22138.1 JAS22720.1 JAS23072.1 JAS25006.1 JAS25768.1 GDIQ01036052 JAN58685.1 IACF01005278 LAB70864.1 GDIP01140921 JAL62793.1 GDIQ01101139 JAL50587.1 PYGN01000050 PSN56140.1 GEDC01029182 GEDC01024486 GEDC01016035 JAS08116.1 JAS12812.1 JAS21263.1 GDIP01169663 JAJ53739.1 ADTU01010985 ADTU01010986 ADTU01010987 GDIQ01248330 JAK03395.1 GDIQ01217481 JAK34244.1 GBHO01004699 JAG38905.1 LRGB01000389 KZS19289.1 GDIQ01035012 JAN59725.1 KQ976540 KYM81185.1 GDIP01210375 JAJ13027.1 ACPB03014183 GDIQ01069489 JAN25248.1 GDIP01062691 JAM41024.1 KZ288187 PBC34719.1 KQ980581 KYN15465.1 GDIQ01084694 JAN10043.1 KZS19290.1 JR038044 AEY58093.1 GDIP01105282 GDIP01097375 JAM06340.1 JR038045 AEY58094.1 KZ288315 PBC28333.1 NEVH01006567 PNF37882.1 GDIP01111181 JAL92533.1 GDIQ01096230 JAL55496.1 LNIX01000021 OXA43787.1 PNF37881.1 GDIP01022054 JAM81661.1 GDIQ01183115 JAK68610.1 GDIP01109537 JAL94177.1 GDIP01131493 JAL72221.1 GDIP01034895 JAM68820.1 GDIQ01009228 JAN85509.1 KQ981673 KYN38170.1 GDIQ01007615 JAN87122.1 GDIP01152383 JAJ71019.1 GBHO01042701 GBHO01003941 JAG00903.1 JAG39663.1 GL888932 EGI57368.1 GDIP01109535 JAL94179.1 GDIP01212074 JAJ11328.1 GDIQ01123725 JAL28001.1 GDIQ01055811 JAN38926.1 GDIP01184280 JAJ39122.1 GDIQ01184049 GDIP01134043 JAK67676.1 JAL69671.1 GDIP01109536 JAL94178.1 GDIP01184281 JAJ39121.1 GDIP01101490 JAM02225.1 GDIP01196765 JAJ26637.1 GDIQ01096228 JAL55498.1 GDIQ01121174 JAL30552.1 GDIP01212073 GDIQ01187448 GDIQ01123724 JAJ11329.1 JAK64277.1 GDIP01069438 JAM34277.1 GDIP01134089 JAL69625.1 GDIP01193179 JAJ30223.1 GDIQ01055810 JAN38927.1 GDIP01111182 JAL92532.1 GDHC01015042 JAQ03587.1 GDIQ01105580 JAL46146.1 KM025027 AIP87047.1 GDIQ01096229 JAL55497.1 GDIQ01121173 JAL30553.1 GDIQ01175304 GDIQ01073580 JAN21157.1 GBYB01000656 JAG70423.1 QOIP01000004 RLU23754.1 GDIQ01077337 JAN17400.1 GDIQ01239538 JAK12187.1 GDIP01181228 JAJ42174.1 GDIQ01082235 JAN12502.1 GDIP01179325 JAJ44077.1
Proteomes
Pfam
PF03098 An_peroxidase
Interpro
SUPFAM
SSF48113
SSF48113
Gene 3D
ProteinModelPortal
A0A194Q1N0
A0A212ERF1
A0A2W1BFY1
A0A2H1WEE0
A0A1B6MDD3
A0A232F238
+ More
K7IYZ2 A0A1B6CUT2 A0A2S2R1E6 A0A2H8TD85 A0A0P4VM15 X1WIY8 A0A146M0F8 T1I7W3 E9IRB0 A0A0J7KKC2 A0A0P5K7G1 E2ADZ2 A0A0P5LUG0 A0A0K8T8R0 A0A0P5M7P6 A0A0P6DES3 A0A1B6DAS3 A0A0N8EE02 A0A2P2IA20 A0A0P5SF47 A0A0N8C9P0 A0A2P8ZI40 A0A1B6CHC4 A0A0P5CJI8 A0A158NBE1 A0A0P5G0R9 A0A0P5I633 A0A0A9ZAS3 A0A162Q098 A0A0N8EED2 A0A195BAD8 A0A0P4ZMN0 T1HAC7 A0A0P6EWY1 A0A0P5Y961 A0A2A3EU99 A0A195DRC5 A0A0P6E070 A0A162Q0A9 V9IAE9 A0A0N8CUK9 V9ICS7 A0A2A3E974 A0A2J7RAN0 A0A0P5ULQ4 A0A0P5RU22 A0A226DGH2 A0A088AE09 A0A2J7RAL9 A0A0P6AZR4 A0A0N8BFE3 A0A0P5URI8 A0A0P5TIN6 A0A0N8DGW9 A0A0N8ENK5 A0A195FC13 A0A0P6JHY0 A0A0P5DSD1 A0A0A9Z8J0 F4X8U9 A0A0N8CQ89 A0A0P5AI81 A0A0P5PVA2 A0A0P6FQD9 A0A0P5BHY2 A0A0P5KKX2 A0A0P5UR53 A0A0N8A457 A0A0P5VCX6 A0A0P5AMC4 A0A0P5SG58 A0A0P5Q175 A0A0N7ZU83 A0A088AE08 A0A0P5XPB9 A0A0P5U773 A0A0P5AWG5 A0A0P6G880 A0A0P5ULU2 A0A146L8Z9 A0A0P5SEE6 A0A089FQD2 A0A0P5RV48 A0A0P5QGH8 A0A0N8E0L1 A0A0C9R1B3 A0A3L8DTP5 A0A0P6DHJ3 A0A0P5GUL0 A0A0P5BQS1 A0A0P6D9I9 A0A0P5BVJ2
K7IYZ2 A0A1B6CUT2 A0A2S2R1E6 A0A2H8TD85 A0A0P4VM15 X1WIY8 A0A146M0F8 T1I7W3 E9IRB0 A0A0J7KKC2 A0A0P5K7G1 E2ADZ2 A0A0P5LUG0 A0A0K8T8R0 A0A0P5M7P6 A0A0P6DES3 A0A1B6DAS3 A0A0N8EE02 A0A2P2IA20 A0A0P5SF47 A0A0N8C9P0 A0A2P8ZI40 A0A1B6CHC4 A0A0P5CJI8 A0A158NBE1 A0A0P5G0R9 A0A0P5I633 A0A0A9ZAS3 A0A162Q098 A0A0N8EED2 A0A195BAD8 A0A0P4ZMN0 T1HAC7 A0A0P6EWY1 A0A0P5Y961 A0A2A3EU99 A0A195DRC5 A0A0P6E070 A0A162Q0A9 V9IAE9 A0A0N8CUK9 V9ICS7 A0A2A3E974 A0A2J7RAN0 A0A0P5ULQ4 A0A0P5RU22 A0A226DGH2 A0A088AE09 A0A2J7RAL9 A0A0P6AZR4 A0A0N8BFE3 A0A0P5URI8 A0A0P5TIN6 A0A0N8DGW9 A0A0N8ENK5 A0A195FC13 A0A0P6JHY0 A0A0P5DSD1 A0A0A9Z8J0 F4X8U9 A0A0N8CQ89 A0A0P5AI81 A0A0P5PVA2 A0A0P6FQD9 A0A0P5BHY2 A0A0P5KKX2 A0A0P5UR53 A0A0N8A457 A0A0P5VCX6 A0A0P5AMC4 A0A0P5SG58 A0A0P5Q175 A0A0N7ZU83 A0A088AE08 A0A0P5XPB9 A0A0P5U773 A0A0P5AWG5 A0A0P6G880 A0A0P5ULU2 A0A146L8Z9 A0A0P5SEE6 A0A089FQD2 A0A0P5RV48 A0A0P5QGH8 A0A0N8E0L1 A0A0C9R1B3 A0A3L8DTP5 A0A0P6DHJ3 A0A0P5GUL0 A0A0P5BQS1 A0A0P6D9I9 A0A0P5BVJ2
PDB
3R5Q
E-value=7.86596e-65,
Score=628
Ontologies
GO
PANTHER
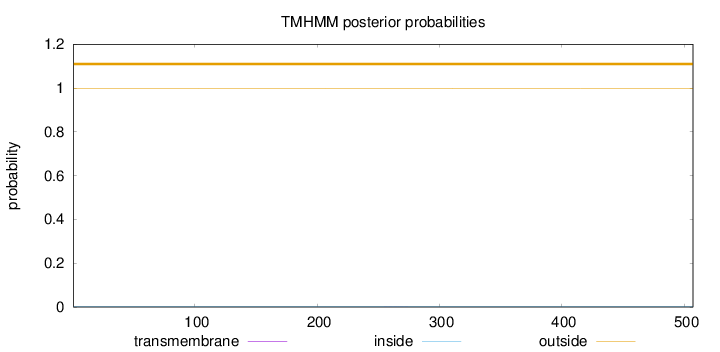
Topology
Length:
507
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00518999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00229
outside
1 - 507
Population Genetic Test Statistics
Pi
227.306615
Theta
163.204341
Tajima's D
1.461289
CLR
0.28587
CSRT
0.778811059447028
Interpretation
Uncertain
