Gene
KWMTBOMO03267
Pre Gene Modal
BGIBMGA013607
Annotation
PREDICTED:_uncharacterized_protein_LOC101737056_[Bombyx_mori]
Full name
Fatty acid 2-hydroxylase
Location in the cell
PlasmaMembrane Reliability : 2.977
Sequence
CDS
ATGAAAATAATTCGTGTTTCATTTCAGAGCCGTCTAGACTGGTCGAAGCCTTTGCTGTGCCAGCTCCACGCCATAGCGCCGGACTACGAGAAATGGGTTAACAGCGCGGTCTACCGGAAATGTAGACTTTTCGCGAGCCCATTGCTCGAGAGTCTAACATATACGCCATGGTATTTCGTTCCTATGTTCTGGATACCTGTAATTTTATATCTTGGCTGGACGCAGTATTACGAGCAAGTCGCTTGTGGAGAACATTGCACAGATGCGAACATCACAACATTCCAGTACGTGTATTATATTTCATTGGGAGTGCTGTTGTGGACGATACTGGAATACTCGCTGCATCGGTGGATCTTTCACTTGGACCCGGGGTCGTCGCTCATCATGATCAAACTGCATTTCTTAATACACGGCATGCACCATAAAGTTCCCTTTGATGGTTTGCGCCAGGTGTTCCCCCCCATACCGGCTCTAGGTATAACCACAATAATATACTCAGCACTTCGGCCGTTTCTGCCCTACCCTGTGCTAGTGCTGACCGGAGGACTCATTGGCTACCTAACTTACGACATGATACATTACTATGTTCATCACGGCTCGCCAAAAGACGGAACCTATCTGTACGCGATGAAGAGATACCACTCGAACCATCATTTCCTCAATCACGACAAAGCATACGGCATAAGCTCGAAAATGTGGGACCATGTGTTCAGGACAGTTGCTCACGTAAAAAAATTAAGTTTTAGCTTACGTTGGTAG
Protein
MKIIRVSFQSRLDWSKPLLCQLHAIAPDYEKWVNSAVYRKCRLFASPLLESLTYTPWYFVPMFWIPVILYLGWTQYYEQVACGEHCTDANITTFQYVYYISLGVLLWTILEYSLHRWIFHLDPGSSLIMIKLHFLIHGMHHKVPFDGLRQVFPPIPALGITTIIYSALRPFLPYPVLVLTGGLIGYLTYDMIHYYVHHGSPKDGTYLYAMKRYHSNHHFLNHDKAYGISSKMWDHVFRTVAHVKKLSFSLRW
Summary
Description
Catalyzes stereospecific hydroxylation of free fatty acids at the C-2 position to produce (R)-2-hydroxy fatty acids, which are building blocks of sphingolipids and glycosphingolipids common in neural tissue and epidermis. Plays an essential role in the synthesis of galactosphingolipids of the myelin sheath. Responsible for the synthesis of sphingolipids and glycosphingolipids involved in the formation of epidermal lamellar bodies critical for skin permeability barrier. Participates in the synthesis of glycosphingolipids and a fraction of type II wax diesters in sebaceous gland, specifically regulating hair follicle homeostasis. Involved in the synthesis of sphingolipids of plasma membrane rafts, controlling lipid raft mobility and trafficking of raft-associated proteins.
Cofactor
Fe cation
Zn(2+)
Zn(2+)
Similarity
Belongs to the sterol desaturase family.
Belongs to the sterol desaturase family. SCS7 subfamily.
Belongs to the cytochrome b5 family.
Belongs to the sterol desaturase family. SCS7 subfamily.
Belongs to the cytochrome b5 family.
Uniprot
H9JVP4
A0A2H1WEE9
A0A2A4JL10
A0A194Q1K1
A0A2W1BMW4
A0A0N1IFR2
+ More
U4U9U4 J3JVT4 N6TSI4 A0A182VDR4 A0A182WYS7 A0A182TQS2 A0A182LHF8 A0A182Y6V3 V5I951 A0A182HYA7 A0A1Q3FNK4 A0A182RL09 A0A1Q3FPA2 A0A182KEA3 A0A1Q3FPT5 A0A182NT21 A0A182Q052 A0A182P0Q9 A0A182WFR1 A0A084VD31 A0A182IRX2 A0A182HBU5 A0A182M5D7 A0A182FLM1 A0A1S4F5S1 Q17EC2 W5JVL3 A0A2M4BSA7 A0A067RBL4 A0A1Y1KCD3 B4KND0 D6WPH5 A0A139WEB0 B4LMP7 J9JXJ3 B4J6P4 A0A2S2P225 A0A2S2QVV5 A0A0M8ZX84 A0A2A3EE22 A0A0K8ULM3 B4MR65 A0A1B0GH23 A0A3B0JSG5 A0A1B6DBJ9 B3MDQ4 B5E0J8 A0A087ZYY0 A0A2J7PSJ3 T1DSV2 A0A3Q0J8C9 A0A034W3A7 A0A0L7RGV0 A0A1L8DYG7 A0A034W2R9 A0A1I8M2E7 A0A3Q0J3B8 A0A1L8DYW2 A0A1B0AGG5 A0A0L0BVZ3 A0A1I8PZ47 B0W2P1 A0A1S3EWE3 A0A1B6IXI3 A0A2K6ER96 A0A091VKL8 A0A1B0G1H6 A0A091MQH8 A0A091PX90 A0A1A9WR42 A0A1B6GPF9 A0A1A9XFG5 A0A087QND3 F1S436 H0VSY7 A0A1J1HMU2 A0A310SW65 A0A091IFQ9 A0A1B0BAP2 A0A2Y9G4G0 A0A2P4SK20 A0A091FP89 A0A093PTC5 A0A091GNH1 B4GBP4 G5B5X0 A0A1A9V3A1 L5MC40 A0A226MG67 A0A154PQC5 A0A340XUH1 A0A3L7I082 A0A3L8SGG7
U4U9U4 J3JVT4 N6TSI4 A0A182VDR4 A0A182WYS7 A0A182TQS2 A0A182LHF8 A0A182Y6V3 V5I951 A0A182HYA7 A0A1Q3FNK4 A0A182RL09 A0A1Q3FPA2 A0A182KEA3 A0A1Q3FPT5 A0A182NT21 A0A182Q052 A0A182P0Q9 A0A182WFR1 A0A084VD31 A0A182IRX2 A0A182HBU5 A0A182M5D7 A0A182FLM1 A0A1S4F5S1 Q17EC2 W5JVL3 A0A2M4BSA7 A0A067RBL4 A0A1Y1KCD3 B4KND0 D6WPH5 A0A139WEB0 B4LMP7 J9JXJ3 B4J6P4 A0A2S2P225 A0A2S2QVV5 A0A0M8ZX84 A0A2A3EE22 A0A0K8ULM3 B4MR65 A0A1B0GH23 A0A3B0JSG5 A0A1B6DBJ9 B3MDQ4 B5E0J8 A0A087ZYY0 A0A2J7PSJ3 T1DSV2 A0A3Q0J8C9 A0A034W3A7 A0A0L7RGV0 A0A1L8DYG7 A0A034W2R9 A0A1I8M2E7 A0A3Q0J3B8 A0A1L8DYW2 A0A1B0AGG5 A0A0L0BVZ3 A0A1I8PZ47 B0W2P1 A0A1S3EWE3 A0A1B6IXI3 A0A2K6ER96 A0A091VKL8 A0A1B0G1H6 A0A091MQH8 A0A091PX90 A0A1A9WR42 A0A1B6GPF9 A0A1A9XFG5 A0A087QND3 F1S436 H0VSY7 A0A1J1HMU2 A0A310SW65 A0A091IFQ9 A0A1B0BAP2 A0A2Y9G4G0 A0A2P4SK20 A0A091FP89 A0A093PTC5 A0A091GNH1 B4GBP4 G5B5X0 A0A1A9V3A1 L5MC40 A0A226MG67 A0A154PQC5 A0A340XUH1 A0A3L7I082 A0A3L8SGG7
EC Number
1.-.-.-
Pubmed
EMBL
BABH01032275
BABH01032276
BABH01032277
BABH01032278
BABH01032279
BABH01032280
+ More
BABH01032281 BABH01032282 BABH01032283 ODYU01008112 SOQ51431.1 NWSH01001169 PCG72274.1 KQ459579 KPI99457.1 KZ150071 PZC74060.1 KQ460650 KPJ13123.1 KB632149 ERL89118.1 BT127352 AEE62314.1 APGK01020738 APGK01020739 KB740234 ENN81003.1 GALX01003545 JAB64921.1 APCN01004511 GFDL01005982 JAV29063.1 GFDL01005614 JAV29431.1 GFDL01005559 JAV29486.1 AXCN02001792 ATLV01011135 KE524643 KFB35875.1 JXUM01032434 KQ560955 KXJ80223.1 AXCM01002891 CH477283 EAT44857.1 ADMH02000297 ETN67075.1 GGFJ01006825 MBW55966.1 KK852611 KDR20253.1 GEZM01088641 GEZM01088640 GEZM01088639 JAV58051.1 CH933808 EDW09983.2 KQ971354 EFA06812.1 KYB26252.1 CH940648 EDW60988.1 KRF79721.1 ABLF02018471 ABLF02018472 ABLF02018473 ABLF02018474 CH916367 EDW00947.1 GGMR01010347 MBY22966.1 GGMS01012059 MBY81262.1 KQ435809 KOX72965.1 KZ288269 PBC29977.1 GDHF01024700 GDHF01009000 GDHF01005657 GDHF01003629 GDHF01000784 JAI27614.1 JAI43314.1 JAI46657.1 JAI48685.1 JAI51530.1 CH963849 EDW74604.1 AJWK01003896 AJWK01003897 OUUW01000001 SPP75671.1 GEDC01014221 JAS23077.1 CH902619 EDV36439.1 CM000071 EDY68997.2 KRT02095.1 NEVH01021928 PNF19304.1 GAMD01000065 JAB01526.1 GAKP01010337 JAC48615.1 KQ414596 KOC70085.1 GFDF01002618 JAV11466.1 GAKP01010340 GAKP01010335 GAKP01010333 JAC48617.1 GFDF01002619 JAV11465.1 JRES01001254 KNC24188.1 DS231827 EDS29349.1 GECU01016051 JAS91655.1 KL411055 KFR03013.1 CCAG010005392 KK835055 KFP79397.1 KK679659 KFQ11883.1 GECZ01005526 JAS64243.1 KL225750 KFM02737.1 AEMK02000039 AAKN02026378 AAKN02026379 CVRI01000011 CRK89267.1 KQ759870 OAD62446.1 KL218516 KFP06200.1 JXJN01011086 JXJN01011087 PPHD01041061 POI24454.1 KK719362 KFO63090.1 KL669996 KFW77475.1 KL448320 KFO82826.1 CH479181 EDW32302.1 JH168604 EHB04681.1 KB101785 ELK36174.1 MCFN01000948 OXB54287.1 KQ435037 KZC14103.1 RAZU01000121 RLQ71628.1 QUSF01000023 RLW01321.1
BABH01032281 BABH01032282 BABH01032283 ODYU01008112 SOQ51431.1 NWSH01001169 PCG72274.1 KQ459579 KPI99457.1 KZ150071 PZC74060.1 KQ460650 KPJ13123.1 KB632149 ERL89118.1 BT127352 AEE62314.1 APGK01020738 APGK01020739 KB740234 ENN81003.1 GALX01003545 JAB64921.1 APCN01004511 GFDL01005982 JAV29063.1 GFDL01005614 JAV29431.1 GFDL01005559 JAV29486.1 AXCN02001792 ATLV01011135 KE524643 KFB35875.1 JXUM01032434 KQ560955 KXJ80223.1 AXCM01002891 CH477283 EAT44857.1 ADMH02000297 ETN67075.1 GGFJ01006825 MBW55966.1 KK852611 KDR20253.1 GEZM01088641 GEZM01088640 GEZM01088639 JAV58051.1 CH933808 EDW09983.2 KQ971354 EFA06812.1 KYB26252.1 CH940648 EDW60988.1 KRF79721.1 ABLF02018471 ABLF02018472 ABLF02018473 ABLF02018474 CH916367 EDW00947.1 GGMR01010347 MBY22966.1 GGMS01012059 MBY81262.1 KQ435809 KOX72965.1 KZ288269 PBC29977.1 GDHF01024700 GDHF01009000 GDHF01005657 GDHF01003629 GDHF01000784 JAI27614.1 JAI43314.1 JAI46657.1 JAI48685.1 JAI51530.1 CH963849 EDW74604.1 AJWK01003896 AJWK01003897 OUUW01000001 SPP75671.1 GEDC01014221 JAS23077.1 CH902619 EDV36439.1 CM000071 EDY68997.2 KRT02095.1 NEVH01021928 PNF19304.1 GAMD01000065 JAB01526.1 GAKP01010337 JAC48615.1 KQ414596 KOC70085.1 GFDF01002618 JAV11466.1 GAKP01010340 GAKP01010335 GAKP01010333 JAC48617.1 GFDF01002619 JAV11465.1 JRES01001254 KNC24188.1 DS231827 EDS29349.1 GECU01016051 JAS91655.1 KL411055 KFR03013.1 CCAG010005392 KK835055 KFP79397.1 KK679659 KFQ11883.1 GECZ01005526 JAS64243.1 KL225750 KFM02737.1 AEMK02000039 AAKN02026378 AAKN02026379 CVRI01000011 CRK89267.1 KQ759870 OAD62446.1 KL218516 KFP06200.1 JXJN01011086 JXJN01011087 PPHD01041061 POI24454.1 KK719362 KFO63090.1 KL669996 KFW77475.1 KL448320 KFO82826.1 CH479181 EDW32302.1 JH168604 EHB04681.1 KB101785 ELK36174.1 MCFN01000948 OXB54287.1 KQ435037 KZC14103.1 RAZU01000121 RLQ71628.1 QUSF01000023 RLW01321.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000030742
UP000019118
+ More
UP000075903 UP000076407 UP000075902 UP000075882 UP000076408 UP000075840 UP000075900 UP000075881 UP000075884 UP000075886 UP000075885 UP000075920 UP000030765 UP000075880 UP000069940 UP000249989 UP000075883 UP000069272 UP000008820 UP000000673 UP000027135 UP000009192 UP000007266 UP000008792 UP000007819 UP000001070 UP000053105 UP000242457 UP000007798 UP000092461 UP000268350 UP000007801 UP000001819 UP000005203 UP000235965 UP000079169 UP000053825 UP000095301 UP000092445 UP000037069 UP000095300 UP000002320 UP000081671 UP000233160 UP000053283 UP000092444 UP000091820 UP000092443 UP000053286 UP000008227 UP000005447 UP000183832 UP000054308 UP000092460 UP000248480 UP000052976 UP000053258 UP000053760 UP000008744 UP000006813 UP000078200 UP000198323 UP000076502 UP000265300 UP000273346 UP000276834
UP000075903 UP000076407 UP000075902 UP000075882 UP000076408 UP000075840 UP000075900 UP000075881 UP000075884 UP000075886 UP000075885 UP000075920 UP000030765 UP000075880 UP000069940 UP000249989 UP000075883 UP000069272 UP000008820 UP000000673 UP000027135 UP000009192 UP000007266 UP000008792 UP000007819 UP000001070 UP000053105 UP000242457 UP000007798 UP000092461 UP000268350 UP000007801 UP000001819 UP000005203 UP000235965 UP000079169 UP000053825 UP000095301 UP000092445 UP000037069 UP000095300 UP000002320 UP000081671 UP000233160 UP000053283 UP000092444 UP000091820 UP000092443 UP000053286 UP000008227 UP000005447 UP000183832 UP000054308 UP000092460 UP000248480 UP000052976 UP000053258 UP000053760 UP000008744 UP000006813 UP000078200 UP000198323 UP000076502 UP000265300 UP000273346 UP000276834
Interpro
IPR012337
RNaseH-like_sf
+ More
IPR008906 HATC_C_dom
IPR006694 Fatty_acid_hydroxylase
IPR010255 Haem_peroxidase_sf
IPR019791 Haem_peroxidase_animal
IPR037120 Haem_peroxidase_sf_animal
IPR036400 Cyt_B5-like_heme/steroid_sf
IPR001199 Cyt_B5-like_heme/steroid-bd
IPR018506 Cyt_B5_heme-BS
IPR029590 Pxt
IPR014430 Scs7
IPR008906 HATC_C_dom
IPR006694 Fatty_acid_hydroxylase
IPR010255 Haem_peroxidase_sf
IPR019791 Haem_peroxidase_animal
IPR037120 Haem_peroxidase_sf_animal
IPR036400 Cyt_B5-like_heme/steroid_sf
IPR001199 Cyt_B5-like_heme/steroid-bd
IPR018506 Cyt_B5_heme-BS
IPR029590 Pxt
IPR014430 Scs7
Gene 3D
ProteinModelPortal
H9JVP4
A0A2H1WEE9
A0A2A4JL10
A0A194Q1K1
A0A2W1BMW4
A0A0N1IFR2
+ More
U4U9U4 J3JVT4 N6TSI4 A0A182VDR4 A0A182WYS7 A0A182TQS2 A0A182LHF8 A0A182Y6V3 V5I951 A0A182HYA7 A0A1Q3FNK4 A0A182RL09 A0A1Q3FPA2 A0A182KEA3 A0A1Q3FPT5 A0A182NT21 A0A182Q052 A0A182P0Q9 A0A182WFR1 A0A084VD31 A0A182IRX2 A0A182HBU5 A0A182M5D7 A0A182FLM1 A0A1S4F5S1 Q17EC2 W5JVL3 A0A2M4BSA7 A0A067RBL4 A0A1Y1KCD3 B4KND0 D6WPH5 A0A139WEB0 B4LMP7 J9JXJ3 B4J6P4 A0A2S2P225 A0A2S2QVV5 A0A0M8ZX84 A0A2A3EE22 A0A0K8ULM3 B4MR65 A0A1B0GH23 A0A3B0JSG5 A0A1B6DBJ9 B3MDQ4 B5E0J8 A0A087ZYY0 A0A2J7PSJ3 T1DSV2 A0A3Q0J8C9 A0A034W3A7 A0A0L7RGV0 A0A1L8DYG7 A0A034W2R9 A0A1I8M2E7 A0A3Q0J3B8 A0A1L8DYW2 A0A1B0AGG5 A0A0L0BVZ3 A0A1I8PZ47 B0W2P1 A0A1S3EWE3 A0A1B6IXI3 A0A2K6ER96 A0A091VKL8 A0A1B0G1H6 A0A091MQH8 A0A091PX90 A0A1A9WR42 A0A1B6GPF9 A0A1A9XFG5 A0A087QND3 F1S436 H0VSY7 A0A1J1HMU2 A0A310SW65 A0A091IFQ9 A0A1B0BAP2 A0A2Y9G4G0 A0A2P4SK20 A0A091FP89 A0A093PTC5 A0A091GNH1 B4GBP4 G5B5X0 A0A1A9V3A1 L5MC40 A0A226MG67 A0A154PQC5 A0A340XUH1 A0A3L7I082 A0A3L8SGG7
U4U9U4 J3JVT4 N6TSI4 A0A182VDR4 A0A182WYS7 A0A182TQS2 A0A182LHF8 A0A182Y6V3 V5I951 A0A182HYA7 A0A1Q3FNK4 A0A182RL09 A0A1Q3FPA2 A0A182KEA3 A0A1Q3FPT5 A0A182NT21 A0A182Q052 A0A182P0Q9 A0A182WFR1 A0A084VD31 A0A182IRX2 A0A182HBU5 A0A182M5D7 A0A182FLM1 A0A1S4F5S1 Q17EC2 W5JVL3 A0A2M4BSA7 A0A067RBL4 A0A1Y1KCD3 B4KND0 D6WPH5 A0A139WEB0 B4LMP7 J9JXJ3 B4J6P4 A0A2S2P225 A0A2S2QVV5 A0A0M8ZX84 A0A2A3EE22 A0A0K8ULM3 B4MR65 A0A1B0GH23 A0A3B0JSG5 A0A1B6DBJ9 B3MDQ4 B5E0J8 A0A087ZYY0 A0A2J7PSJ3 T1DSV2 A0A3Q0J8C9 A0A034W3A7 A0A0L7RGV0 A0A1L8DYG7 A0A034W2R9 A0A1I8M2E7 A0A3Q0J3B8 A0A1L8DYW2 A0A1B0AGG5 A0A0L0BVZ3 A0A1I8PZ47 B0W2P1 A0A1S3EWE3 A0A1B6IXI3 A0A2K6ER96 A0A091VKL8 A0A1B0G1H6 A0A091MQH8 A0A091PX90 A0A1A9WR42 A0A1B6GPF9 A0A1A9XFG5 A0A087QND3 F1S436 H0VSY7 A0A1J1HMU2 A0A310SW65 A0A091IFQ9 A0A1B0BAP2 A0A2Y9G4G0 A0A2P4SK20 A0A091FP89 A0A093PTC5 A0A091GNH1 B4GBP4 G5B5X0 A0A1A9V3A1 L5MC40 A0A226MG67 A0A154PQC5 A0A340XUH1 A0A3L7I082 A0A3L8SGG7
PDB
4ZR0
E-value=9.23468e-28,
Score=305
Ontologies
GO
GO:0008610
GO:0006979
GO:0005506
GO:0020037
GO:0046983
GO:0016021
GO:0004601
GO:0016491
GO:0080132
GO:0006633
GO:0005789
GO:0046872
GO:0006631
GO:0005783
GO:0001949
GO:0044857
GO:0042634
GO:0061436
GO:0042127
GO:0006682
GO:0032287
GO:0030258
GO:0006679
GO:0032286
GO:0004702
GO:0007178
GO:0016020
GO:0005634
GO:0006270
GO:0042555
GO:0048384
GO:0055114
PANTHER
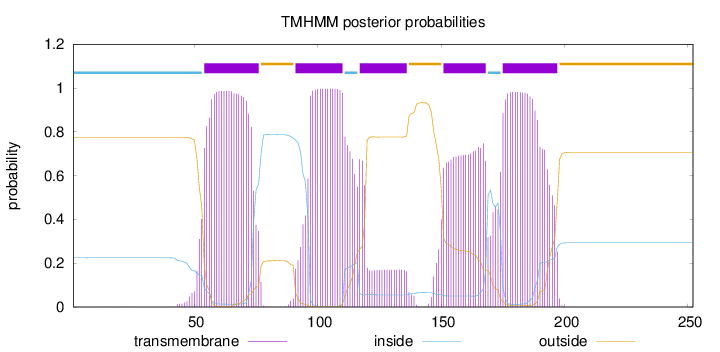
Topology
Subcellular location
Endoplasmic reticulum membrane
Length:
252
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
81.3420400000001
Exp number, first 60 AAs:
7.45533
Total prob of N-in:
0.22462
inside
1 - 53
TMhelix
54 - 76
outside
77 - 90
TMhelix
91 - 110
inside
111 - 116
TMhelix
117 - 136
outside
137 - 150
TMhelix
151 - 168
inside
169 - 174
TMhelix
175 - 197
outside
198 - 252
Population Genetic Test Statistics
Pi
165.534443
Theta
161.02436
Tajima's D
1.133842
CLR
0.279217
CSRT
0.695515224238788
Interpretation
Uncertain
